

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.5
(1017 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
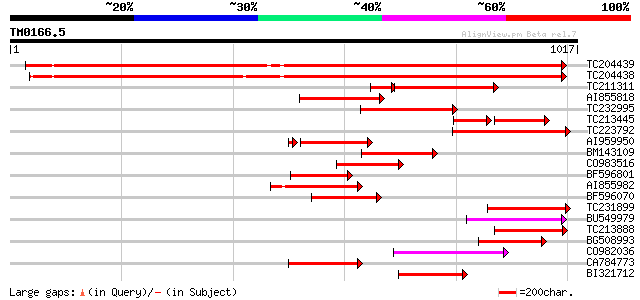
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 914 0.0
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 909 0.0
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 207 4e-62
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 221 2e-57
TC232995 211 1e-54
TC213445 129 6e-49
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 179 7e-45
AI959950 164 3e-40
BM143109 155 8e-38
CO983516 151 1e-36
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 150 3e-36
AI855982 142 1e-33
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 130 2e-30
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 130 4e-30
BU549979 123 4e-28
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 122 6e-28
BG508993 120 2e-27
CO982036 120 2e-27
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 119 9e-27
BI321712 110 3e-24
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 914 bits (2362), Expect = 0.0
Identities = 464/974 (47%), Positives = 648/974 (65%), Gaps = 3/974 (0%)
Frame = +1
Query: 29 KSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVL 88
K + G + +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL
Sbjct: 1795 KIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVL 1971
Query: 89 FSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLP 148
G R + + + S CL S DE IW +R GH LR + ++ +RG+P
Sbjct: 1972 MKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIP 2151
Query: 149 RLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVI 208
LK +C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+
Sbjct: 2152 NLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVV 2331
Query: 209 VDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNS 268
VDD+SR+TWV F+R K ET +F ++Q+E + +RSDHG EFEN F E S
Sbjct: 2332 VDDFSRFTWVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTS 2511
Query: 269 QGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISI 328
+GI+H FS TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +++
Sbjct: 2512 EGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTL 2691
Query: 329 RPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGF 388
R T YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+ +
Sbjct: 2692 RRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAY 2871
Query: 389 RIYNIIHQTVEESIQIRFDGKLGSEKSKLFERF---ADLSIDCSEANQPKNSSEDVAPEA 445
R++N +TV ESI + D + K + E D D +++ + +S+ E+
Sbjct: 2872 RVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDES 3051
Query: 446 EASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIE 505
++ +S ++QK HP+ELIIG+ + V TRS E ++S VS IE
Sbjct: 3052 NINQPDKRSSTRIQKM------HPKELIIGDPNRGVTTRSR----EVEIVSNSCFVSKIE 3201
Query: 506 PKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTR 565
PK+V EAL D+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TR
Sbjct: 3202 PKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITR 3381
Query: 566 NKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYIS 625
NKARLVAQGY+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++
Sbjct: 3382 NKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLN 3561
Query: 626 EEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLF 685
EEVYV+QP GF D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF
Sbjct: 3562 EEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLF 3741
Query: 686 CITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPG 745
+++I QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q
Sbjct: 3742 VKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMED 3921
Query: 746 VTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLT 805
++ Q +Y ++KKF M + + +TP L K+E + V Q LYR MIGSLLYLT
Sbjct: 3922 SIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLT 4101
Query: 806 ASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDAD 865
ASRPDI ++V +CAR+Q++P+ +HLT VKRILKY+ GT++ G+MY S L G+CDAD
Sbjct: 4102 ASRPDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDAD 4281
Query: 866 FAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLE 925
+AG +RKSTSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L+
Sbjct: 4282 WAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLK 4461
Query: 926 DYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQ 985
+Y + + +YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++L++VDT+ Q
Sbjct: 4462 EYNVEQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQ 4641
Query: 986 WADIFTKPLAEDRF 999
ADIFTK L ++F
Sbjct: 4642 IADIFTKALDANQF 4683
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 909 bits (2349), Expect = 0.0
Identities = 462/964 (47%), Positives = 639/964 (65%)
Frame = +1
Query: 36 IGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFSGKRKN 95
+ DG +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL G R
Sbjct: 1822 VHDG-LPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSK 1995
Query: 96 YIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLPRLKYSSE 155
+ + + S CL S DE IW +R GH LR + ++ +RG+P LK
Sbjct: 1996 DNCYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEG 2175
Query: 156 ALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVIVDDYSRW 215
+C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+VDD+SR+
Sbjct: 2176 RICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRF 2355
Query: 216 TWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQGISHNF 275
TWV F+R K +T +F ++Q+E + +RSDHG EFEN F E S+GI+H F
Sbjct: 2356 TWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEF 2535
Query: 276 SCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRPILEKT 335
S TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +++R T
Sbjct: 2536 SAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTT 2715
Query: 336 PYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIH 395
YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+ +R++N
Sbjct: 2716 LYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRT 2895
Query: 396 QTVEESIQIRFDGKLGSEKSKLFERFADLSIDCSEANQPKNSSEDVAPEAEASEAAPTTS 455
+TV ESI + D + K + E D+ S+E+ A++
Sbjct: 2896 RTVMESINVVVDDLTPARKKDVEE---DVRTSGDNVADTAKSAENAENSDSATDEPNINQ 3066
Query: 456 DQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALED 515
+ RI HP+ELIIG+ + V TRS E ++S VS IEPK+V EAL D
Sbjct: 3067 PDKRPSIRIQKMHPKELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEPKNVKEALTD 3234
Query: 516 KGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGY 575
+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TRNKARLVAQGY
Sbjct: 3235 EFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGY 3414
Query: 576 SQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPG 635
+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++EE YV+QP G
Sbjct: 3415 TQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKG 3594
Query: 636 FEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILI 695
F D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF +++I
Sbjct: 3595 FVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMI 3774
Query: 696 VQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYT 755
QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q ++ Q KY
Sbjct: 3775 AQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYA 3954
Query: 756 LELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSV 815
++KKF M + + +TP L K+E + V Q LYR MIGSLLYLTASRPDI ++V
Sbjct: 3955 KNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAV 4134
Query: 816 HLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKS 875
+CAR+Q++P+ +HL VKRILKY+ GT++ G+MY S+ L G+CDAD+AG +RKS
Sbjct: 4135 GVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKS 4314
Query: 876 TSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVP 935
TSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L++Y + +
Sbjct: 4315 TSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMT 4494
Query: 936 IYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLA 995
+YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++LE+VDT+ Q ADIFTK L
Sbjct: 4495 LYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKALD 4674
Query: 996 EDRF 999
++F
Sbjct: 4675 ANQF 4686
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 207 bits (526), Expect(2) = 4e-62
Identities = 106/188 (56%), Positives = 135/188 (71%)
Frame = +3
Query: 690 KNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYI 749
K LI+ IYVDDIIFG+ + +CKEF +LM+ FE SM GELK+ LG+QI Q+ +I
Sbjct: 477 KETFLIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIFI 656
Query: 750 HQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRP 809
HQ+KYT LK+F M + TPMH + I++K+E + K Y GMI SL YLT+SRP
Sbjct: 657 HQEKYTKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTSSRP 836
Query: 810 DILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGD 869
DI+F V LCARFQS P+ +H+TAVKRIL+YL GTTN L ++K SE+ L G+CD FAGD
Sbjct: 837 DIVFVVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYFAGD 1016
Query: 870 RVERKSTS 877
+VERKSTS
Sbjct: 1017KVERKSTS 1040
Score = 51.2 bits (121), Expect(2) = 4e-62
Identities = 26/47 (55%), Positives = 30/47 (63%)
Frame = +2
Query: 647 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDI 693
K +YGLKQA RAWYERLSSFL+ N F RG D LF K ++
Sbjct: 347 KTLSCVYGLKQALRAWYERLSSFLVSNGFTRGITDPALFRKAQKGNL 487
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 221 bits (562), Expect = 2e-57
Identities = 104/153 (67%), Positives = 129/153 (83%)
Frame = -3
Query: 520 LGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQE 579
+ MQEEL+QF +N+VW L+ KP+ + VIGTKWVFRNKL+E G + RNKARLVA+GY+Q+E
Sbjct: 461 IAMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEE 282
Query: 580 GIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDD 639
GIDY ET++PVARLE IR+L+++ N L+QMDVKSAFLNG I EEVYV+QPPGFE
Sbjct: 281 GIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIP 102
Query: 640 KYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQ 672
P HVYKL+K+LYGLKQAPRAWYER+S+FLL+
Sbjct: 101 DKPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>TC232995
Length = 1009
Score = 211 bits (537), Expect = 1e-54
Identities = 107/173 (61%), Positives = 125/173 (71%)
Frame = +2
Query: 630 VKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITY 689
V+QPPGFE P+HVYKL+K+LYGLKQAPRAWYERLS+FLL+ EF RGK D TLF
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 690 KNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYI 749
NDIL+VQIYVDDIIFGS N SLCKEFS MQ+EFEMSMMGELKYFLG+QI Q +I
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 750 HQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLL 802
+Q KY EL+K+F M TPM C L+K+E + K YR IG ++
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVV 520
>TC213445
Length = 705
Score = 129 bits (325), Expect(2) = 6e-49
Identities = 57/98 (58%), Positives = 78/98 (79%)
Frame = +1
Query: 870 RVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGL 929
+ +R+STS +CHF+GS LV+W SK+QN++ LSTAEAEY+SA + Q WM+ L DYGL
Sbjct: 400 KTDRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARSYYAQIFWMRQQLFDYGL 579
Query: 930 SLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIR 967
L +PI CDNT+AI+LSKN IL+SR KHIE+++H++R
Sbjct: 580 KLDHIPIRCDNTSAINLSKNHILYSRTKHIEIRHHFLR 693
Score = 84.3 bits (207), Expect(2) = 6e-49
Identities = 38/68 (55%), Positives = 51/68 (74%)
Frame = +2
Query: 797 MIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEY 856
MI S LYL+ SRP I+FSV +C R+Q++P+E+HL+ +KRI++YL G NLGL Y K S Y
Sbjct: 197 MIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPKNSSY 376
Query: 857 TLSGFCDA 864
L G+ DA
Sbjct: 377 NLVGYSDA 400
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein, partial
(7%)
Length = 804
Score = 179 bits (453), Expect = 7e-45
Identities = 96/217 (44%), Positives = 137/217 (62%), Gaps = 4/217 (1%)
Frame = +1
Query: 794 YRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMY--- 850
+R +IGSL YL SRP+I F+V L +RF PR +H+ A KR+L+ +KGT G+++
Sbjct: 22 FRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVLFPFK 201
Query: 851 RKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSA 910
K+ + L G+ D+D+ D + KST G V SSK+Q+ IALST EAEYV+A
Sbjct: 202 AKSGKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAEYVAA 381
Query: 911 ATCCTQTIWMKNHLEDYGLSLKK-VPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDH 969
+ Q +WM N LE+ L +K V + DN +AI+L+K+P LH R+KHIE+++HYIRD
Sbjct: 382 SLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHYIRDQ 561
Query: 970 VQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENL 1006
V KG +++EY + Q AD+ TKP+ RF I L
Sbjct: 562 VSKGNVTVEYCKAEEQLADLMTKPIQVSRFKQICSEL 672
>AI959950
Length = 466
Score = 164 bits (415), Expect(2) = 3e-40
Identities = 82/129 (63%), Positives = 101/129 (77%)
Frame = -1
Query: 522 MQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGI 581
MQEELDQF KN+V L+ PK V+G KW+F NKL+E G+V R KARLVA+GYSQQEGI
Sbjct: 388 MQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEGI 209
Query: 582 DYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKY 641
DY +TF+ VARLE I +L+SF+ N+ L+QMDVKSAFLNG I +EVYV+QPPGFE++
Sbjct: 208 DYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFENETL 29
Query: 642 PDHVYKLKK 650
HV+KL K
Sbjct: 28 HQHVFKLNK 2
Score = 20.8 bits (42), Expect(2) = 3e-40
Identities = 7/16 (43%), Positives = 13/16 (80%)
Frame = -2
Query: 500 LVSLIEPKSVDEALED 515
L+ ++PK +DEA++D
Sbjct: 453 LIFEMKPKHIDEAIKD 406
>BM143109
Length = 415
Score = 155 bits (392), Expect = 8e-38
Identities = 80/135 (59%), Positives = 96/135 (70%)
Frame = +1
Query: 632 QPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKN 691
QPP ++ + P+HV+KLKK LYGLKQA RAWYE LS FLL F +GK D LF N
Sbjct: 4 QPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLN 183
Query: 692 DILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQ 751
DIL+VQIYVDDIIFGS N SLCK+FS+ MQ EFEMSMM EL +FLG+QI Q +I Q
Sbjct: 184 DILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFISQ 363
Query: 752 KKYTLELLKKFNMSD 766
KY +L+ +F M +
Sbjct: 364 SKYCKDLIHRFGMEN 408
>CO983516
Length = 724
Score = 151 bits (382), Expect = 1e-36
Identities = 74/120 (61%), Positives = 91/120 (75%)
Frame = +2
Query: 587 FSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVY 646
F PVARLE+IRLL+ + L+QMDVKSAFLNGY++EEVYV+QP GF D +PDHVY
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 647 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFG 706
+LKK+LYGLKQAPRAWYERL+ L Q + +G D TLF +++I QIYVDDI+FG
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFG 724
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 150 bits (379), Expect = 3e-36
Identities = 69/111 (62%), Positives = 92/111 (82%)
Frame = +3
Query: 505 EPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVT 564
EPK++ EA+ D WI+ MQEEL+QF +N+VW L+ KP+ + VIGTKWVFRNKL+E G +
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 565 RNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDV 615
RNKARLVA+GY+Q+EGIDY ET++PVARLEAIR+L++++ N L+QMDV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>AI855982
Length = 484
Score = 142 bits (357), Expect = 1e-33
Identities = 77/165 (46%), Positives = 106/165 (63%)
Frame = +2
Query: 469 PEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQ 528
P + IIG+ V TR LK L + VS+IEPK++ EA+ D WI+ MQEEL+Q
Sbjct: 2 PLDNIIGDISKGVTTRHSLKD----LCNNMAFVSMIEPKNIKEAIVDDNWIIAMQEELNQ 169
Query: 529 FTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFS 588
F +N+VW L+ KP + VI TKWVFRNKL+E + +KARLVA+GY+Q +G+DY T++
Sbjct: 170 FERNNVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIIIHKARLVAEGYNQVDGLDYEHTYA 349
Query: 589 PVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQP 633
+ARL I + +S+ N TL+ SA L+G + EVYV QP
Sbjct: 350 SIARL*VIIMPLSYVYIMNSTLYHYACVSALLHGLLLHEVYVDQP 484
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 130 bits (328), Expect = 2e-30
Identities = 63/127 (49%), Positives = 89/127 (69%)
Frame = -2
Query: 541 PKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLI 600
P G +G +WV+ K+ GEV R KARLVA+GY+Q GIDY +TFSPVA+L +RL +
Sbjct: 397 PPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVRLFL 218
Query: 601 SFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPR 660
+ + + LHQ+D+K+AFL+G + E++Y++QPPGF V KL +SLYGLKQ+PR
Sbjct: 217 AMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQSPR 38
Query: 661 AWYERLS 667
AW+ + S
Sbjct: 37 AWFGKFS 17
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment), partial
(30%)
Length = 687
Score = 130 bits (326), Expect = 4e-30
Identities = 61/150 (40%), Positives = 95/150 (62%), Gaps = 1/150 (0%)
Frame = +2
Query: 858 LSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQT 917
LSG+CDAD+AG ++R+STSG C F+G NLV+W SK+Q +A S+AEAEY S A +
Sbjct: 17 LSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCEL 196
Query: 918 IWMKNHLEDYGLSLK-KVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLS 976
+W+K L++ + ++ +YCDN AA+ ++ NP+ H R KHIE+ H+IR+ + +
Sbjct: 197 MWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSKEIV 376
Query: 977 LEYVDTDHQWADIFTKPLAEDRFLFILENL 1006
E++ ++ Q DI TK L + + L
Sbjct: 377 TEFIGSNDQPVDILTKSLRGPKIQIVCSKL 466
>BU549979
Length = 615
Score = 123 bits (309), Expect = 4e-28
Identities = 63/183 (34%), Positives = 107/183 (58%), Gaps = 3/183 (1%)
Frame = -1
Query: 820 RFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGS 879
R+QS+P H K++++YL+GT + LMY++T+ + G+ D+DFAG R+STSG
Sbjct: 606 RYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRSTSGY 427
Query: 880 CHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGL---SLKKVPI 936
L +V+W S +Q IA ST E E+V + +W+K+ + + + + +
Sbjct: 426 IFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRPLKL 247
Query: 937 YCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAE 996
YCDN AA+ ++KN +R+KHI++KY IR+ V++ + +E+V+T+ D TK +
Sbjct: 246 YCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTELMIVDPLTKGMTP 67
Query: 997 DRF 999
F
Sbjct: 66 KNF 58
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 122 bits (307), Expect = 6e-28
Identities = 57/132 (43%), Positives = 89/132 (67%), Gaps = 1/132 (0%)
Frame = +3
Query: 870 RVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGL 929
R +RKST+G F+G TW SK+Q + LST EAEYV+A +C IW++N L++ +
Sbjct: 6 RDDRKSTTGFVFFMGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELKM 185
Query: 930 SLKKVPIYC-DNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWAD 988
++ C DN +A++L+KNP+ H ++KHI+ +YH+IR+ ++K + L+YV + Q AD
Sbjct: 186 PQEEPMEICVDNKSALALAKNPVFHEKSKHIDTRYHFIRECIEKKEVKLKYVMSQDQAAD 365
Query: 989 IFTKPLAEDRFL 1000
IFTKPL + F+
Sbjct: 366 IFTKPLKLETFV 401
>BG508993
Length = 374
Score = 120 bits (302), Expect = 2e-27
Identities = 58/123 (47%), Positives = 80/123 (64%), Gaps = 1/123 (0%)
Frame = +1
Query: 841 KGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIAL 900
KGT + GL Y ++ Y L GFCD+DFAGD +RKST+G F+G + TWSSK+Q + L
Sbjct: 4 KGTIDFGLFYSPSNNYKLVGFCDSDFAGDVDDRKSTTGFVFFMGDCVFTWSSKKQGIVTL 183
Query: 901 STAEAEYVSAATCCTQTIWMKNHLEDYGLSLKK-VPIYCDNTAAISLSKNPILHSRAKHI 959
T EAEYV+A +C IW++ LE+ L K+ IY DN +A L+KN + H R+KHI
Sbjct: 184 FTCEAEYVAATSCTCHAIWLRRLLEELQLLQKESTKIYVDNRSAQELAKNSVFHERSKHI 363
Query: 960 EVK 962
+ +
Sbjct: 364 DTR 372
>CO982036
Length = 674
Score = 120 bits (302), Expect = 2e-27
Identities = 72/212 (33%), Positives = 117/212 (54%), Gaps = 5/212 (2%)
Frame = -2
Query: 689 YKNDILIVQ--IYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGV 746
YK IL V +YVD II GS+ +L + + + + F + ++G+L YF+ I++ P +
Sbjct: 673 YKTHILTVYLLVYVDIIITGSSC-TLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDL 497
Query: 747 TYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTA 806
+ + E+ + +PM TC L K + YR ++G+L Y T
Sbjct: 496 LF-SLRTSIFEIFCRKPR*QAQPISSPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTV 320
Query: 807 SRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYR---KTSEYTLSGFCD 863
RP+I F+V+ +F S+P ++H T VKRIL+YLKG+ + GL + + + GFCD
Sbjct: 319 IRPEISFAVNKVCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCD 140
Query: 864 ADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQ 895
AD+A +++STSG+ FLG NL++W +Q
Sbjct: 139 ADWASAVDDKRSTSGAAVFLGPNLISWWXXKQ 44
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 119 bits (297), Expect = 9e-27
Identities = 56/134 (41%), Positives = 85/134 (62%)
Frame = +3
Query: 500 LVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNE 559
L SL P ++ EAL+ GW M +E+ N W L+ P G +G +WV+ K+
Sbjct: 3 LSSLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGP 182
Query: 560 KGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAF 619
G+V R KARLVA+GY+Q GI+Y +TFSPV L +RL ++ + + LHQ+D+K+AF
Sbjct: 183 NGKVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAF 362
Query: 620 LNGYISEEVYVKQP 633
L+G + E++Y++QP
Sbjct: 363 LHGDLEEDIYMEQP 404
>BI321712
Length = 399
Score = 110 bits (275), Expect = 3e-24
Identities = 58/124 (46%), Positives = 77/124 (61%)
Frame = -3
Query: 698 IYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLE 757
+YVDD+IF NPS+ +EF K M EFEM+ MG + Y+LGI++ Q +I Q+ Y E
Sbjct: 379 LYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQEDKGIFITQEGYAKE 200
Query: 758 LLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHL 817
+LKKF M D N TPM L K E V LY+ +IGSL YLT +RPDIL+ V +
Sbjct: 199 VLKKFKMDDANPVGTPMECGSKLSKHEKGENVDPTLYKSLIGSLRYLTCTRPDILYVVGV 20
Query: 818 CARF 821
+R+
Sbjct: 19 VSRY 8
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,959,724
Number of Sequences: 63676
Number of extensions: 631306
Number of successful extensions: 3188
Number of sequences better than 10.0: 130
Number of HSP's better than 10.0 without gapping: 3103
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3143
length of query: 1017
length of database: 12,639,632
effective HSP length: 107
effective length of query: 910
effective length of database: 5,826,300
effective search space: 5301933000
effective search space used: 5301933000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0166.5


