

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.4
(1003 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
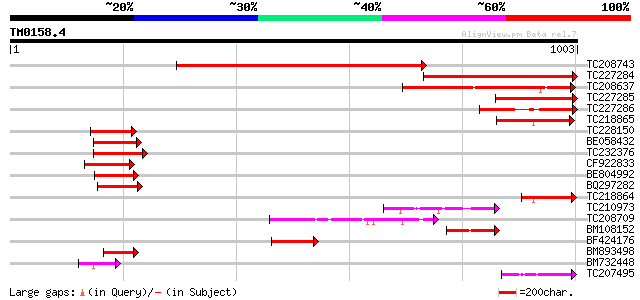
Score E
Sequences producing significant alignments: (bits) Value
TC208743 similar to UP|Q9SMX9 (Q9SMX9) Squamosa promoter binding... 778 0.0
TC227284 similar to UP|O82651 (O82651) Squamosa-promoter binding... 472 e-133
TC208637 similar to UP|O82651 (O82651) Squamosa-promoter binding... 284 1e-76
TC227285 similar to UP|O82651 (O82651) Squamosa-promoter binding... 255 8e-68
TC227286 similar to UP|O82651 (O82651) Squamosa-promoter binding... 226 3e-59
TC218865 similar to UP|O82651 (O82651) Squamosa-promoter binding... 143 3e-34
TC228150 homologue to UP|Q9S849 (Q9S849) Squamosa promoter bindi... 124 2e-28
BE058432 similar to SP|Q38741|SBP1_ Squamosa-promoter binding pr... 124 2e-28
TC232376 123 4e-28
CF922833 120 2e-27
BE804992 similar to PIR|T52297|T522 squamosa promoter binding pr... 111 1e-24
BQ297282 similar to PIR|T52607|T526 squamosa promoter binding pr... 105 8e-23
TC218864 similar to UP|O82651 (O82651) Squamosa-promoter binding... 104 2e-22
TC210973 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fr... 104 2e-22
TC208709 similar to UP|Q8S9G8 (Q8S9G8) AT5g18830/F17K4_80, parti... 104 2e-22
BM108152 101 2e-21
BF424176 weakly similar to PIR|T52601|T526 squamosa promoter bin... 94 4e-19
BM893498 similar to GP|18650611|gb| At1g27370/F17L21_16 {Arabido... 88 2e-17
BM732448 similar to SP|O04003|LG1_M LIGULELESS1 protein. [Maize]... 83 5e-16
TC207495 weakly similar to UP|Q9SMW9 (Q9SMW9) SPL1-Related3 prot... 73 5e-13
>TC208743 similar to UP|Q9SMX9 (Q9SMX9) Squamosa promoter binding
protein-like 1, partial (35%)
Length = 1326
Score = 778 bits (2010), Expect = 0.0
Identities = 382/441 (86%), Positives = 409/441 (92%)
Frame = +3
Query: 296 NLLREGGSSRESEMVPTLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMS 355
NLLRE GSSR+SEM+ TL SNGSQGSPTD RQH+TVS+ KMQQ+V+HAHDARA DQQ+ S
Sbjct: 3 NLLREDGSSRKSEMMSTLFSNGSQGSPTDTRQHETVSIAKMQQQVMHAHDARAADQQITS 182
Query: 356 YIKPSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHVTSSLD 415
IKPS+SN+PPAY+EARDSTAGQIKMNNFDLNDIYIDSDDG+EDLERLP +TN VTSSLD
Sbjct: 183 SIKPSMSNSPPAYSEARDSTAGQIKMNNFDLNDIYIDSDDGMEDLERLPVSTNLVTSSLD 362
Query: 416 YPWTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRA 475
YPW QQDSHQSSPPQTS NS+S SA SPSS +GEAQ RTDRIV KLFGKEP+DFPL+LRA
Sbjct: 363 YPWAQQDSHQSSPPQTSGNSDSASAQSPSSFSGEAQSRTDRIVFKLFGKEPNDFPLVLRA 542
Query: 476 QILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRT 535
QILDWLSHSPTD+ESYIRPGCIVLTIYLRQAEA+WEE+CYDLTSSL RLLDVSDDTFWR
Sbjct: 543 QILDWLSHSPTDMESYIRPGCIVLTIYLRQAEALWEELCYDLTSSLNRLLDVSDDTFWRN 722
Query: 536 GWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIR 595
GWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKR QFSVKGVN+IR
Sbjct: 723 GWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRAQFSVKGVNLIR 902
Query: 596 PATRLMCALEGKYLVCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSS 655
PATRLMCALEGKYLVCEDAH SMDQ S+E D L C+QFSC+VPV NGRGFIEIEDQGLSS
Sbjct: 903 PATRLMCALEGKYLVCEDAHMSMDQSSKEPDELQCVQFSCSVPVMNGRGFIEIEDQGLSS 1082
Query: 656 SFFPFIVAEEDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQL 715
SFFPFIV EEDVCS+IC LEPLLELSETDPDIEGTGKIKAK+QAMDFIHE+GWLLH SQL
Sbjct: 1083SFFPFIVVEEDVCSEICTLEPLLELSETDPDIEGTGKIKAKNQAMDFIHEMGWLLHRSQL 1262
Query: 716 KSRMVHLSSSTDLFPLKRFKW 736
K RMV L+SS DLFPLKRFKW
Sbjct: 1263KLRMVQLNSSEDLFPLKRFKW 1325
>TC227284 similar to UP|O82651 (O82651) Squamosa-promoter binding protein-like
1, partial (18%)
Length = 1017
Score = 472 bits (1215), Expect = e-133
Identities = 230/275 (83%), Positives = 252/275 (91%), Gaps = 3/275 (1%)
Frame = +1
Query: 732 KRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDLALSEMGLLHRAVRRNSKQL 791
KRFKWL+EFSMDHDWCA V+KLLNLLL+GTV+TGDHPSL LALSEMGLLH+AVRRNSKQL
Sbjct: 1 KRFKWLIEFSMDHDWCAAVRKLLNLLLDGTVNTGDHPSLYLALSEMGLLHKAVRRNSKQL 180
Query: 792 VELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLD 851
VE LL YVPENIS+++ PE KALV+GE Q+FLFRPD G AGLTPLHIAAGKDGSEDVLD
Sbjct: 181 VEWLLRYVPENISDKLGPEDKALVDGENQTFLFRPDVDGTAGLTPLHIAAGKDGSEDVLD 360
Query: 852 ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPT 911
ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHY YIHLVQKKINK+QGA HVVVEIP+
Sbjct: 361 ALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYAYIHLVQKKINKKQGAAHVVVEIPS 540
Query: 912 NLTENDTNQKQNESSTTFEIAK---TRDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVA 968
N+TEN+TN+KQNE ST FEI K R QGHCKLCD ++SCRTAVGRS+VY+PAMLSMVA
Sbjct: 541 NMTENNTNKKQNELSTIFEIGKPEVRRGQGHCKLCDNRISCRTAVGRSMVYRPAMLSMVA 720
Query: 969 VAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
+AAVCVCVALLFKSSPEV+ MFRPF WE+LDFGTS
Sbjct: 721 IAAVCVCVALLFKSSPEVICMFRPFRWENLDFGTS 825
>TC208637 similar to UP|O82651 (O82651) Squamosa-promoter binding protein-like
1, partial (8%)
Length = 1303
Score = 284 bits (727), Expect = 1e-76
Identities = 153/316 (48%), Positives = 207/316 (65%), Gaps = 10/316 (3%)
Frame = +3
Query: 696 KSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLN 755
K+QA+DF+ E+GWLLH S +K ++ ++ DLF RF WL++FSMDH WCAV+KKLL+
Sbjct: 3 KTQALDFLQEMGWLLHRSHVKFKLGSMAPFHDLFQFNRFAWLVDFSMDHGWCAVMKKLLD 182
Query: 756 LLLEGTVSTGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALV 815
++ EG V G+H S++LAL MGLLHRAV+RN + +VELLL +VP S+ E K +
Sbjct: 183 IIFEGGVDAGEHASIELALLNMGLLHRAVKRNCRPMVELLLRFVPVKTSDGADSEMKQVA 362
Query: 816 EGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDAL-TNDPCMVGI-EAWKNARDSTG 873
E + FLFRPD GPAGLTPLH+AA GSE + DP G K ARDSTG
Sbjct: 363 EAPDR-FLFRPDTVGPAGLTPLHVAASMSGSETCVGMH*PYDPSNGGN**HGKCARDSTG 539
Query: 874 STPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEIAK 933
TP D+A LRG+Y+YI LVQ K NK+ H +V+IP + +T QKQ++ + T +
Sbjct: 540 LTPNDHACLRGYYSYIQLVQNKTNKKGERQH-LVDIPGTVVFFNTTQKQSDGNRTCRVPS 716
Query: 934 TRDQ--------GHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPE 985
+ + C+ C K++ + ++VY+P MLSMV +A VCVCVALLFKSSP
Sbjct: 717 LKTEKIETTAMPRQCRACQQKVA-YGGMKTAMVYRPVMLSMVTIAVVCVCVALLFKSSPR 893
Query: 986 VLYMFRPFSWESLDFG 1001
V Y+F+PF+WESL++G
Sbjct: 894 VYYVFQPFNWESLEYG 941
>TC227285 similar to UP|O82651 (O82651) Squamosa-promoter binding protein-like
1, partial (10%)
Length = 628
Score = 255 bits (651), Expect = 8e-68
Identities = 124/147 (84%), Positives = 134/147 (90%), Gaps = 3/147 (2%)
Frame = +1
Query: 860 VGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTN 919
VGIEAWKNARDSTGSTPEDYARLRGHY YIHLVQKKINKRQGA HVVVEIP+N TE++TN
Sbjct: 43 VGIEAWKNARDSTGSTPEDYARLRGHYAYIHLVQKKINKRQGAAHVVVEIPSNTTESNTN 222
Query: 920 QKQNESSTTFEIAKT---RDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCV 976
+KQNE STTFEI K R QGHCKLCD ++SCRTAVGRSLVY+PAMLSMVA+AAVCVCV
Sbjct: 223 EKQNELSTTFEIGKAEVIRGQGHCKLCDKRISCRTAVGRSLVYRPAMLSMVAIAAVCVCV 402
Query: 977 ALLFKSSPEVLYMFRPFSWESLDFGTS 1003
ALLFKSSPEV+ MFRPF WE+LDFGTS
Sbjct: 403 ALLFKSSPEVICMFRPFRWENLDFGTS 483
>TC227286 similar to UP|O82651 (O82651) Squamosa-promoter binding protein-like
1, partial (13%)
Length = 721
Score = 226 bits (577), Expect = 3e-59
Identities = 119/172 (69%), Positives = 129/172 (74%)
Frame = +3
Query: 832 AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHL 891
AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHY YIHL
Sbjct: 3 AGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYAYIHL 182
Query: 892 VQKKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQGHCKLCDIKLSCRT 951
VQKK N ++ SS+ KLCD ++SCRT
Sbjct: 183 VQKK-----------------------N*QETRSSSC-------GG*DSKLCDNRISCRT 272
Query: 952 AVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
AVGRS+VY+PAMLSMVA+AAVCVCVALLFKSSPEV+ MFRPF WE+LDFGTS
Sbjct: 273 AVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVICMFRPFRWENLDFGTS 428
>TC218865 similar to UP|O82651 (O82651) Squamosa-promoter binding
protein-like 1, partial (7%)
Length = 457
Score = 143 bits (361), Expect = 3e-34
Identities = 78/146 (53%), Positives = 102/146 (69%), Gaps = 8/146 (5%)
Frame = +3
Query: 861 GIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTNQ 920
G EAWK+A+D+TG TP DYA +RG+Y+YI LVQ K + + HV+ +IP L +++T Q
Sbjct: 24 GTEAWKSAQDATGLTPYDYASMRGYYSYIQLVQSKTSNTCKSQHVL-DIPGTLVDSNTKQ 200
Query: 921 KQNE--------SSTTFEIAKTRDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAV 972
KQ++ S T +I T C LC KL+ + R+LVY+PAMLSMVA+AAV
Sbjct: 201 KQSDRHRSSKVSSLQTEKIETTAMPRRCGLCQQKLAYG-GMRRALVYRPAMLSMVAIAAV 377
Query: 973 CVCVALLFKSSPEVLYMFRPFSWESL 998
CVCVALLFKSSP+V Y+F+PFSWESL
Sbjct: 378 CVCVALLFKSSPKVYYVFQPFSWESL 455
>TC228150 homologue to UP|Q9S849 (Q9S849) Squamosa promoter binding
protein-like 8, partial (23%)
Length = 685
Score = 124 bits (312), Expect = 2e-28
Identities = 56/81 (69%), Positives = 62/81 (76%)
Frame = +1
Query: 143 ASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEF 202
+SN CQ E C ADLS+AK YHRRHKVCE HSKA + QRFCQQCSRFH+L EF
Sbjct: 49 SSNSPRCQAEGCNADLSQAKHYHRRHKVCEFHSKAATVIAAGLTQRFCQQCSRFHLLSEF 228
Query: 203 DEGKRSCRRRLAGHNKRRRKT 223
D GKRSCR+RLA HN+RRRKT
Sbjct: 229 DNGKRSCRKRLADHNRRRRKT 291
>BE058432 similar to SP|Q38741|SBP1_ Squamosa-promoter binding protein 1.
[Garden snapdragon] {Antirrhinum majus}, partial (58%)
Length = 433
Score = 124 bits (312), Expect = 2e-28
Identities = 59/84 (70%), Positives = 64/84 (75%)
Frame = +1
Query: 149 CQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRS 208
CQ E C ADL+ AK YHRRHKVCE HSKA +V QRFCQQCSRFH L EFDE KRS
Sbjct: 22 CQAERCGADLTDAKRYHRRHKVCEFHSKAPVVVVAGLRQRFCQQCSRFHDLAEFDESKRS 201
Query: 209 CRRRLAGHNKRRRKTNNEAVPNGN 232
CRRRLAGHN+RRRK+N EA G+
Sbjct: 202 CRRRLAGHNERRRKSNPEASNEGS 273
>TC232376
Length = 1033
Score = 123 bits (309), Expect = 4e-28
Identities = 55/96 (57%), Positives = 67/96 (69%)
Frame = +2
Query: 149 CQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRS 208
C V+ C ADLS +DYHRRHKVCE+HSK + +G QRFCQQCSRFH L+EFDEGKRS
Sbjct: 41 CLVDGCHADLSNCRDYHRRHKVCEVHSKTAQVSIGGQKQRFCQQCSRFHSLEEFDEGKRS 220
Query: 209 CRRRLAGHNKRRRKTNNEAVPNGNPLNDDQTSSYLL 244
CR+RL GHN+RRRK E++ + + LL
Sbjct: 221 CRKRLDGHNRRRRKPQPESLTRSGSFLSNYQGTQLL 328
>CF922833
Length = 664
Score = 120 bits (302), Expect = 2e-27
Identities = 57/90 (63%), Positives = 67/90 (74%)
Frame = +2
Query: 132 NGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQ 191
+GK+S GGG+ + CQV++C ADLS AK YHRRHKVCE H+KA + QRFCQ
Sbjct: 203 HGKRSGSKGGGSMPPS-CQVDNCDADLSEAKQYHRRHKVCEYHAKAPSVHMAGLQQRFCQ 379
Query: 192 QCSRFHMLQEFDEGKRSCRRRLAGHNKRRR 221
QCSRFH L EFD+ KRSCR RLAGHN+RRR
Sbjct: 380 QCSRFHELSEFDDSKRSCRTRLAGHNERRR 469
>BE804992 similar to PIR|T52297|T522 squamosa promoter binding
protein-homolog 5 [imported] - garden snapdragon
(fragment), partial (22%)
Length = 418
Score = 111 bits (278), Expect = 1e-24
Identities = 50/78 (64%), Positives = 63/78 (80%)
Frame = +2
Query: 151 VEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCR 210
VE C L AK+YHRRH+VC+ HSKA +A+V QRFCQQCSRFH++ EFD+ KRSCR
Sbjct: 2 VEGCHVALVNAKEYHRRHRVCDKHSKAPKAVVLGLEQRFCQQCSRFHVVSEFDDSKRSCR 181
Query: 211 RRLAGHNKRRRKTNNEAV 228
RRLAGHN+RRRK+++ +V
Sbjct: 182 RRLAGHNERRRKSSHLSV 235
>BQ297282 similar to PIR|T52607|T526 squamosa promoter binding protein 5
[imported] - Arabidopsis thaliana, partial (42%)
Length = 427
Score = 105 bits (263), Expect = 8e-23
Identities = 49/80 (61%), Positives = 57/80 (71%)
Frame = +2
Query: 156 ADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAG 215
ADL AK YHRRH+VCE H KA LV QRFCQQCSRFH L EFD+ KRSCR LAG
Sbjct: 170 ADLHEAKQYHRRHRVCEYHVKAQVVLVDEVRQRFCQQCSRFHELAEFDDTKRSCRSSLAG 349
Query: 216 HNKRRRKTNNEAVPNGNPLN 235
HN+RRRK ++++ G+ N
Sbjct: 350 HNERRRKNSDQSQAEGSSRN 409
>TC218864 similar to UP|O82651 (O82651) Squamosa-promoter binding protein-like
1, partial (7%)
Length = 821
Score = 104 bits (260), Expect = 2e-22
Identities = 55/104 (52%), Positives = 74/104 (70%), Gaps = 7/104 (6%)
Frame = +3
Query: 906 VVEIPTNLTENDTNQKQNES-------STTFEIAKTRDQGHCKLCDIKLSCRTAVGRSLV 958
V++IP NL +++T QKQ++ S E +T HC LC KL + R+LV
Sbjct: 39 VLDIPGNLVDSNTKQKQSDGHRSSKVLSLQTEKIETTAMRHCGLCQQKL-VYGGMRRALV 215
Query: 959 YKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
++PAMLSMVA+AAVCVCVALLFKSSP+V Y+F+PFSWESL++G+
Sbjct: 216 FRPAMLSMVAIAAVCVCVALLFKSSPKVYYVFQPFSWESLEYGS 347
>TC210973 similar to UP|Q9SMW6 (Q9SMW6) Spl1-Related2 protein (Fragment),
partial (15%)
Length = 585
Score = 104 bits (260), Expect = 2e-22
Identities = 69/217 (31%), Positives = 121/217 (54%), Gaps = 12/217 (5%)
Frame = +1
Query: 662 VAEEDVCSDICGLEPLLELSETDPDIEGT------GKIKAKSQAMDFIHEIGWLLHGSQL 715
+A+E +C ++ LE + E D G+ K++ +A+ F++E+GWL +
Sbjct: 1 IADETICKELKPLESEFDEEEKICDAISEEHEHHFGRPKSREEALHFLNELGWLFQRERF 180
Query: 716 KSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLL----LEGT-VSTGDHPSL 770
VH + L RFK+++ F+++ + C +VK LL++L L+G +STG S+
Sbjct: 181 S--YVH---EVPYYSLDRFKFVLTFAVERNCCMLVKTLLDVLVGKHLQGEWLSTG---SV 336
Query: 771 DLALSEMGLLHRAVRRNSKQLVELLLTY-VPENISNEVRPEGKALVEGEKQSFLFRPDAA 829
++ L+ + LL+RAV+ +V+LL+ Y +P G + ++F P+
Sbjct: 337 EM-LNAIQLLNRAVKGKYVGMVDLLIHYSIPSK-------------NGTSRKYVFPPNLE 474
Query: 830 GPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWK 866
GP G+TPLH+AAG GSE V+D+LT+DP +G++ W+
Sbjct: 475 GPGGITPLHLAAGTSGSESVVDSLTSDPQEIGLKCWE 585
>TC208709 similar to UP|Q8S9G8 (Q8S9G8) AT5g18830/F17K4_80, partial (18%)
Length = 1680
Score = 104 bits (259), Expect = 2e-22
Identities = 84/321 (26%), Positives = 155/321 (48%), Gaps = 22/321 (6%)
Frame = +3
Query: 460 KLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTS 519
KL+ P++FP LR QI WL+ P ++E YIRPGC +LTI++ +W + D
Sbjct: 3 KLYDWNPAEFPRRLRHQIFQWLASMPVELEGYIRPGCTILTIFIAMPNIMWINLLKDPLE 182
Query: 520 SLGRLLDVSDDTFWR-TGWVHIRVQHQMAF--IFNGQVVIDTSLPFRSNNYSKILTVSPI 576
+ ++ R T VH+ + M F + +G V + + + K+ V P
Sbjct: 183 YVHDIVAPGKMLSGRGTALVHL---NDMIFRVMKDGTSVTNVKVNMHA---PKLHYVHPT 344
Query: 577 AAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCEDAHESMDQYSEELDGLHCI----Q 632
A K ++F G N+++P RL+ + GKYL CE S ++E D + C
Sbjct: 345 YFEAGKPMEFVACGSNLLQPKFRLLVSFSGKYLKCEYCVPSPHSWTE--DNISCAFDNQL 518
Query: 633 FSCAVPVTN----GRGFIEIEDQGLSSSFFPFIVAEEDVCSDICGLEPLLELSETDPDIE 688
+ VP T G FIE+E++ S+F P ++ ++ +C+++ L+ L++S
Sbjct: 519 YKIYVPHTEESLFGPAFIEVENESGLSNFIPVLIGDKKICTEMKTLQQKLDVSLLSKQFR 698
Query: 689 GT--GKI--------KAKSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKWLM 738
G I + + + D + +I WLL + ++ +++S ++R+ L+
Sbjct: 699 SASGGSICSSCETFALSHTSSSDLLVDIAWLLKDTTSENFDRVMTAS----QIQRYCHLL 866
Query: 739 EFSMDHDWCAVVKKLL-NLLL 758
+F + +D ++ K+L NL++
Sbjct: 867 DFLICNDSTIILGKILPNLII 929
>BM108152
Length = 540
Score = 101 bits (251), Expect = 2e-21
Identities = 52/94 (55%), Positives = 64/94 (67%)
Frame = +1
Query: 773 ALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPA 832
AL EMGLLH+AV+RN + +VE+LL +VP S+ K V F+FRPD GP
Sbjct: 7 ALLEMGLLHKAVKRNCRPMVEILLKFVPVKASDGGDSNEKQ-VNKSPDKFIFRPDTVGPV 183
Query: 833 GLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWK 866
GLTPLH+AA GS +VLDALT+DP MVG EAW+
Sbjct: 184 GLTPLHVAASMHGSXNVLDALTDDPGMVGTEAWE 285
>BF424176 weakly similar to PIR|T52601|T526 squamosa promoter binding protein
1 [imported] - Arabidopsis thaliana, partial (5%)
Length = 283
Score = 93.6 bits (231), Expect = 4e-19
Identities = 50/87 (57%), Positives = 58/87 (66%), Gaps = 3/87 (3%)
Frame = +2
Query: 463 GKEPSDFPL--ILRAQILDW-LSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTS 519
GKEP F L + + LD +S +ESYI PGCIV+TIYLRQAEA+WEE+CYDLT
Sbjct: 23 GKEPK*FVLSVLYSTRFLDVVISRVLLILESYILPGCIVMTIYLRQAEALWEELCYDLTP 202
Query: 520 SLGRLLDVSDDTFWRTGWVHIRVQHQM 546
S L+DVS DT TGW I QHQM
Sbjct: 203 SSNMLMDVSHDTISETGWADIMAQHQM 283
>BM893498 similar to GP|18650611|gb| At1g27370/F17L21_16 {Arabidopsis
thaliana}, partial (14%)
Length = 421
Score = 88.2 bits (217), Expect = 2e-17
Identities = 36/63 (57%), Positives = 49/63 (77%)
Frame = +1
Query: 166 RRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNN 225
R+H+VCE HSK+ + ++ +RFCQQCSRFH L EFD+ KRSCR+RL+ HN RRRKT
Sbjct: 1 RKHRVCEAHSKSPKVVIAGLERRFCQQCSRFHALSEFDDKKRSCRQRLSDHNARRRKTQP 180
Query: 226 EAV 228
+++
Sbjct: 181 DSI 189
>BM732448 similar to SP|O04003|LG1_M LIGULELESS1 protein. [Maize] {Zea mays},
partial (12%)
Length = 356
Score = 83.2 bits (204), Expect = 5e-16
Identities = 42/83 (50%), Positives = 48/83 (57%), Gaps = 8/83 (9%)
Frame = +1
Query: 122 DREVASWDGGNGKKSRVAGGGASNRA--------VCQVEDCCADLSRAKDYHRRHKVCEM 173
D V +GG + + GGG S R CQ E C ADL+ AK YHRRHKVCE+
Sbjct: 103 DESVVGGEGGLAEDEKKKGGGGSGRRGSSGFFPPSCQAEMCGADLTVAKRYHRRHKVCEL 282
Query: 174 HSKATRALVGNAMQRFCQQCSRF 196
HSKA +V QRFCQQCSRF
Sbjct: 283 HSKAPSVMVAGLRQRFCQQCSRF 351
>TC207495 weakly similar to UP|Q9SMW9 (Q9SMW9) SPL1-Related3 protein
(Fragment), partial (7%)
Length = 1413
Score = 73.2 bits (178), Expect = 5e-13
Identities = 47/136 (34%), Positives = 74/136 (53%), Gaps = 3/136 (2%)
Frame = +2
Query: 870 DSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTF 929
D+ G +P YA +R + +Y LV +K+ RQ + V I N E + + + + ++
Sbjct: 14 DANGQSPHAYAMMRNNDSYNALVARKLADRQRG-EISVTI-ANAIEQQSLRVELKQKQSY 187
Query: 930 EIAKTRDQGHCKLC---DIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEV 986
+ R Q C C +I+ + R L+++P + SM+AVAAVCVCV + F+ P V
Sbjct: 188 LVK--RGQSSCAKCANAEIRYNRRVPGSHGLLHRPFIYSMLAVAAVCVCVCVFFRGRPFV 361
Query: 987 LYMFRPFSWESLDFGT 1002
PFSWE+LD+GT
Sbjct: 362 -GSVAPFSWENLDYGT 406
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,348,795
Number of Sequences: 63676
Number of extensions: 660485
Number of successful extensions: 3762
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 3665
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3740
length of query: 1003
length of database: 12,639,632
effective HSP length: 107
effective length of query: 896
effective length of database: 5,826,300
effective search space: 5220364800
effective search space used: 5220364800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0158.4


