

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
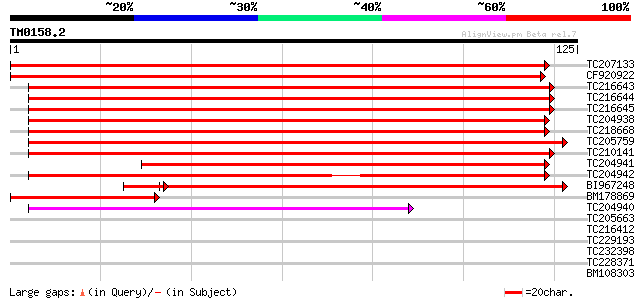
Query= TM0158.2
(125 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207133 similar to GB|AAM70189.1|21636958|AF492760 autophagy AP... 219 3e-58
CF920922 184 1e-47
TC216643 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [... 140 1e-34
TC216644 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [... 139 3e-34
TC216645 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [... 139 3e-34
TC204938 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated ... 132 4e-32
TC218668 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated ... 130 1e-31
TC205759 UP|Q8L5F9 (Q8L5F9) Microtubule associated protein, comp... 126 3e-30
TC210141 homologue to UP|Q8L5F9 (Q8L5F9) Microtubule associated ... 125 5e-30
TC204941 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated ... 112 6e-26
TC204942 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated ... 102 3e-23
BI967248 homologue to GP|21615419|emb microtubule associated pro... 87 1e-18
BM178869 59 4e-10
TC204940 weakly similar to UP|Q6W5F3 (Q6W5F3) Microtubule-associ... 57 3e-09
TC205663 similar to UP|Q6PT61 (Q6PT61) Induced protein MgI1, par... 28 0.82
TC216412 similar to UP|Q41384 (Q41384) Protein kinase (Fragment)... 28 1.4
TC229193 similar to UP|FUM1_ARATH (P93033) Fumarate hydratase 1,... 27 2.4
TC232398 27 2.4
TC228371 similar to UP|Q9AXD7 (Q9AXD7) Response regulator protei... 26 4.1
BM108303 25 9.1
>TC207133 similar to GB|AAM70189.1|21636958|AF492760 autophagy APG8
{Arabidopsis thaliana;} , partial (98%)
Length = 554
Score = 219 bits (558), Expect = 3e-58
Identities = 106/119 (89%), Positives = 114/119 (95%)
Frame = +1
Query: 1 MGGSSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVG 60
MGGSSSF+DE+T +QRL ESR+I+AKYPDRVPVIVERY K DLPEL K+KYLVPRDLSVG
Sbjct: 43 MGGSSSFRDEYTLEQRLGESRDIVAKYPDRVPVIVERYAKCDLPELEKKKYLVPRDLSVG 222
Query: 61 HFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
HFIHILSSRLSLPPGKALFVFVKNTLPQTA+MMDSVYRSF+DEDGFLYMYYSTEKTFGS
Sbjct: 223 HFIHILSSRLSLPPGKALFVFVKNTLPQTANMMDSVYRSFKDEDGFLYMYYSTEKTFGS 399
>CF920922
Length = 544
Score = 184 bits (466), Expect = 1e-47
Identities = 89/118 (75%), Positives = 101/118 (85%)
Frame = -2
Query: 1 MGGSSSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVG 60
MGG+SSF+DE+T +QRL ES +I+AKYPD VPVIVERY K DLPELGK+K LVPRDL VG
Sbjct: 489 MGGNSSFRDEYTLEQRLGESGDIVAKYPDGVPVIVERYGKCDLPELGKKKSLVPRDLFVG 310
Query: 61 HFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
HFIHILSSR LPPGKA FVFVKNTLPQT +MMDS YRSF++ +GF YM+YS KTFG
Sbjct: 309 HFIHILSSRRGLPPGKAFFVFVKNTLPQTVNMMDSGYRSFKEGEGFFYMFYSPGKTFG 136
>TC216643 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (76%)
Length = 717
Score = 140 bits (354), Expect = 1e-34
Identities = 67/116 (57%), Positives = 86/116 (73%)
Frame = +2
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYPDR+PVIVER KSD+PE+ K+KYLVP DL+VG F++
Sbjct: 125 SSFKMEHPLERRQAEASRIREKYPDRIPVIVERAEKSDVPEIDKKKYLVPADLTVGQFVY 304
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L P KA+F+FVKN LP TA+MM ++Y +DEDGFLYM YS E TFG V
Sbjct: 305 VVRKRIKLSPEKAIFIFVKNILPPTAAMMSAIYEENKDEDGFLYMTYSGENTFGLV 472
>TC216644 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (76%)
Length = 740
Score = 139 bits (350), Expect = 3e-34
Identities = 66/116 (56%), Positives = 86/116 (73%)
Frame = +3
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYP+R+PVIVER KSD+PE+ K+KYLVP DL+VG F++
Sbjct: 108 SSFKMEHPLERRQAEASRIREKYPERIPVIVERAEKSDVPEIDKKKYLVPADLTVGQFVY 287
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L P KA+F+FVKN LP TA+MM ++Y +DEDGFLYM YS E TFG V
Sbjct: 288 VVRKRIKLSPEKAIFIFVKNILPPTAAMMSAIYEENKDEDGFLYMTYSGENTFGLV 455
>TC216645 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (76%)
Length = 640
Score = 139 bits (350), Expect = 3e-34
Identities = 66/116 (56%), Positives = 86/116 (73%)
Frame = +1
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SSFK E ++R E+ I KYP+R+PVIVER KSD+PE+ K+KYLVP DL+VG F++
Sbjct: 151 SSFKMEHPLERRQAEASRIREKYPERIPVIVERAEKSDVPEIDKKKYLVPADLTVGQFVY 330
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L P KA+F+FVKN LP TA+MM ++Y +DEDGFLYM YS E TFG V
Sbjct: 331 VVRKRIKLSPEKAIFIFVKNILPPTAAMMSAIYEENKDEDGFLYMTYSGENTFGLV 498
>TC204938 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated protein 1
light chain 3, complete
Length = 655
Score = 132 bits (332), Expect = 4e-32
Identities = 62/115 (53%), Positives = 84/115 (72%)
Frame = +3
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R ES I KYPDR+PVIVE+ +SD+P++ K+KYLVP DL+VG F++
Sbjct: 108 SPFKLEHPLERRQAESARIRDKYPDRIPVIVEKAERSDIPDIDKKKYLVPADLTVGQFVY 287
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
++ R+ + KA+FVFV NTLP TA++M S+Y +D+DGFLYM YS E TFGS
Sbjct: 288 VVRKRIKVSAEKAIFVFVNNTLPPTAALMSSIYEENKDDDGFLYMTYSGENTFGS 452
>TC218668 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated protein 1
light chain 3, complete
Length = 717
Score = 130 bits (328), Expect = 1e-31
Identities = 59/115 (51%), Positives = 85/115 (73%)
Frame = +3
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
+SFK + ++R E+ I KYPDR+PVIVE+ +SD+P++ K+KYLVP DL+VG F++
Sbjct: 258 TSFKLQHPLERRQAEASRIREKYPDRIPVIVEKAERSDIPDIDKKKYLVPADLTVGQFVY 437
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
++ R+ L KA+FVF+ NTLP TA++M ++Y +D+DGFLYM YS E TFGS
Sbjct: 438 VVRKRIKLSAEKAIFVFINNTLPPTAALMSAIYEENKDQDGFLYMTYSGENTFGS 602
>TC205759 UP|Q8L5F9 (Q8L5F9) Microtubule associated protein, complete
Length = 627
Score = 126 bits (316), Expect = 3e-30
Identities = 58/119 (48%), Positives = 83/119 (69%)
Frame = +3
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R E+ I KYPDR+PVIVE+ +SD+P + K+KYLVP DL+VG F++
Sbjct: 93 SYFKQEHDLEKRRAEAARIREKYPDRIPVIVEKAERSDIPSIDKKKYLVPADLTVGQFVY 272
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSVHNI 123
++ R+ L KA+F+FV N LP T ++M ++Y +DEDGFLY+ YS E TFG + +I
Sbjct: 273 VIRKRIKLSAEKAIFIFVDNVLPPTGAIMSAIYDEKKDEDGFLYVTYSGENTFGDLISI 449
>TC210141 homologue to UP|Q8L5F9 (Q8L5F9) Microtubule associated protein,
complete
Length = 605
Score = 125 bits (314), Expect = 5e-30
Identities = 57/116 (49%), Positives = 81/116 (69%)
Frame = +1
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R E+ I KYPDR+PVIVE+ +SD+P + K+KYLVP DL+VG F++
Sbjct: 85 SYFKQEHDLEKRRAEAARIREKYPDRIPVIVEKAERSDIPSIDKKKYLVPADLTVGQFVY 264
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L KA+F+FV N LP T ++M ++Y +DEDGFLY+ YS E TFG +
Sbjct: 265 VIRKRIKLSAEKAIFIFVDNVLPPTGAIMSAIYDEKKDEDGFLYVTYSGENTFGDL 432
>TC204941 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated protein 1
light chain 3, partial (76%)
Length = 465
Score = 112 bits (279), Expect = 6e-26
Identities = 49/90 (54%), Positives = 70/90 (77%)
Frame = +1
Query: 30 RVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQT 89
R+PVIVE+ +SD+P++ K+KYLVP DL+VG F++++ R+ + KA+FVF+ NTLP T
Sbjct: 1 RIPVIVEKAERSDIPDIDKKKYLVPADLTVGQFVYVVRKRIKVSAEKAIFVFIYNTLPPT 180
Query: 90 ASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
A++M S+Y +D+DGFLYM YS E TFGS
Sbjct: 181 AALMSSIYEENKDDDGFLYMTYSGENTFGS 270
>TC204942 homologue to UP|Q6W5F3 (Q6W5F3) Microtubule-associated protein 1
light chain 3, partial (84%)
Length = 1000
Score = 102 bits (255), Expect = 3e-23
Identities = 51/115 (44%), Positives = 75/115 (64%)
Frame = +3
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R ES I KYPDR+PVIVE+ +SD+P++ K+KYLVP DL+VG F++
Sbjct: 528 SPFKLEHPLERRQAESARIRDKYPDRIPVIVEKAERSDIPDIDKKKYLVPADLTVGQFVY 707
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGS 119
++ R+ ++++ A++M S+Y +D+DGFLYM YS E TFGS
Sbjct: 708 VVRKRIK------FWLYL*TESLVAAALMSSIYEENKDDDGFLYMTYSGENTFGS 854
>BI967248 homologue to GP|21615419|emb microtubule associated protein {Cicer
arietinum}, partial (81%)
Length = 446
Score = 87.0 bits (214), Expect(2) = 1e-18
Identities = 37/90 (41%), Positives = 60/90 (66%)
Frame = -3
Query: 34 IVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMM 93
+ E++ +SD+P + K+K LVP +V ++++ R+ L KA+F+FV N LP T ++M
Sbjct: 381 LCEKFXRSDMPSIDKKKCLVPAGFTVAQLVYVIRKRIKLSAEKAIFIFVDNVLPPTGAIM 202
Query: 94 DSVYRSFRDEDGFLYMYYSTEKTFGSVHNI 123
++Y +DEDGFLY+ YS E TFG + +I
Sbjct: 201 SAIYDEKKDEDGFLYVTYSGENTFGDLISI 112
Score = 21.2 bits (43), Expect(2) = 1e-18
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = -2
Query: 26 KYPDRVPVIV 35
K PDR+PVIV
Sbjct: 406 KCPDRIPVIV 377
>BM178869
Length = 420
Score = 59.3 bits (142), Expect = 4e-10
Identities = 26/33 (78%), Positives = 31/33 (93%)
Frame = +2
Query: 1 MGGSSSFKDEFTFDQRLEESREIIAKYPDRVPV 33
MGGSSSF+DE+T +QRL ESR+I+AKYPDRVPV
Sbjct: 32 MGGSSSFRDEYTLEQRLGESRDIVAKYPDRVPV 130
>TC204940 weakly similar to UP|Q6W5F3 (Q6W5F3) Microtubule-associated
protein 1 light chain 3, partial (73%)
Length = 268
Score = 56.6 bits (135), Expect = 3e-09
Identities = 33/85 (38%), Positives = 48/85 (55%)
Frame = +2
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R ES I KY +R+PVIVE+ SD+P++ K+K +V D + G
Sbjct: 8 SPFKLEHPLERRQAESARIRDKYAERIPVIVEKAETSDIPDIDKKKDVVAADWTSGQCGC 187
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQT 89
+ R+ + KA+ V NTLP T
Sbjct: 188VGRRRIXVSAKKAMCEGVSNTLPXT 262
>TC205663 similar to UP|Q6PT61 (Q6PT61) Induced protein MgI1, partial (60%)
Length = 1519
Score = 28.5 bits (62), Expect = 0.82
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = -2
Query: 10 EFTFDQRLEESREIIAKYPDRVPV 33
EF DQRL E REI+ + R+P+
Sbjct: 276 EFRRDQRLRERREIVGSWETRLPL 205
>TC216412 similar to UP|Q41384 (Q41384) Protein kinase (Fragment) , partial
(20%)
Length = 897
Score = 27.7 bits (60), Expect = 1.4
Identities = 16/43 (37%), Positives = 23/43 (53%)
Frame = +1
Query: 44 PELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTL 86
P L ++YLV D V H H+L S+L + + +KNTL
Sbjct: 49 PRL*SKQYLVKEDHEVNHHQHLLQSQLH---NQTHLLELKNTL 168
>TC229193 similar to UP|FUM1_ARATH (P93033) Fumarate hydratase 1,
mitochondrial precursor (Fumarase 1) , partial (40%)
Length = 888
Score = 26.9 bits (58), Expect = 2.4
Identities = 17/46 (36%), Positives = 23/46 (49%), Gaps = 2/46 (4%)
Frame = -2
Query: 52 LVPRDLSVGHFIHILSSRLSLPPGKAL--FVFVKNTLPQTASMMDS 95
+V DL H + R++LP FVF KN LP+TA+ S
Sbjct: 227 MVSHDLCTHHRQSLTLGRVNLPRHNTASWFVFRKNELPKTAAWTTS 90
>TC232398
Length = 1054
Score = 26.9 bits (58), Expect = 2.4
Identities = 16/59 (27%), Positives = 24/59 (40%)
Frame = -2
Query: 67 SSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSVHNINC 125
++ PP + +F + +TL Q E GFL + + TF VHN C
Sbjct: 510 NTEFKCPPTRLIFCSISSTLKQV-------------ECGFLVFHTHLQVTFPHVHNTLC 373
>TC228371 similar to UP|Q9AXD7 (Q9AXD7) Response regulator protein, partial
(53%)
Length = 1068
Score = 26.2 bits (56), Expect = 4.1
Identities = 11/34 (32%), Positives = 21/34 (61%)
Frame = -1
Query: 14 DQRLEESREIIAKYPDRVPVIVERYLKSDLPELG 47
++ + E+ ++IA+ VPVI+ R + + ELG
Sbjct: 996 NENITENSKLIAREYQSVPVILSRLIGESITELG 895
>BM108303
Length = 608
Score = 25.0 bits (53), Expect = 9.1
Identities = 9/16 (56%), Positives = 13/16 (81%)
Frame = -3
Query: 107 LYMYYSTEKTFGSVHN 122
L+++ S+E TFG VHN
Sbjct: 369 LFLHVSSENTFGFVHN 322
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.140 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,807,022
Number of Sequences: 63676
Number of extensions: 55086
Number of successful extensions: 304
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 303
length of query: 125
length of database: 12,639,632
effective HSP length: 86
effective length of query: 39
effective length of database: 7,163,496
effective search space: 279376344
effective search space used: 279376344
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0158.2


