

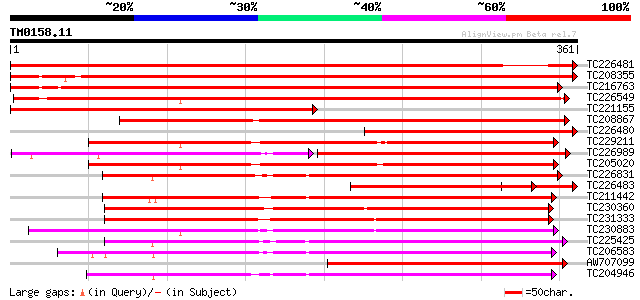
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.11
(361 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226481 similar to UP|Q41328 (Q41328) Pto kinase interactor 1, ... 629 0.0
TC208355 UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, complete 531 e-151
TC216763 UP|Q9LKY3 (Q9LKY3) Pti1 kinase-like protein, complete 529 e-151
TC226549 UP|Q84P43 (Q84P43) Protein kinase Pti1, complete 493 e-140
TC221155 similar to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein ... 345 1e-95
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 261 3e-70
TC226480 similar to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein ... 258 2e-69
TC229211 similar to GB|AAR99874.1|41323411|AY518291 strubbelig r... 246 1e-65
TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 144 1e-65
TC205020 similar to GB|AAR99874.1|41323411|AY518291 strubbelig r... 237 5e-63
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 233 1e-61
TC226483 homologue to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protei... 231 4e-61
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 230 7e-61
TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {A... 229 1e-60
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 229 1e-60
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 220 9e-58
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 219 1e-57
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 217 6e-57
AW707099 weakly similar to GP|3668069|gb| Pto kinase interactor ... 216 1e-56
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 216 2e-56
>TC226481 similar to UP|Q41328 (Q41328) Pto kinase interactor 1, partial
(81%)
Length = 1526
Score = 629 bits (1621), Expect = 0.0
Identities = 316/361 (87%), Positives = 323/361 (88%)
Frame = +2
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCF+CCEEDD HK AESGGPYVVKNP+GNDGN+HAS+TAKQG Q VKPQPIEVPNIS D
Sbjct: 206 MSCFSCCEEDDLHKAAESGGPYVVKNPAGNDGNYHASQTAKQGTQPVKPQPIEVPNISAD 385
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
ELKEVTDNFGQDSLIGEGSYGRVYYGVLK+ AAAIKKLDASKQPDEEFLAQVSMVSRLK
Sbjct: 386 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKSELAAAIKKLDASKQPDEEFLAQVSMVSRLK 565
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
HENFVQLLGY +DG+SRIL YEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA
Sbjct: 566 HENFVQLLGYCIDGSSRILAYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 745
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF
Sbjct: 746 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 925
Query: 241 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV
Sbjct: 926 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 1105
Query: 301 RQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAGETA 360
RQCVDTRLGGEYPP KALQPLLTARPGPAGET
Sbjct: 1106RQCVDTRLGGEYPP----------------------------KALQPLLTARPGPAGETP 1201
Query: 361 N 361
N
Sbjct: 1202N 1204
>TC208355 UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, complete
Length = 1536
Score = 531 bits (1369), Expect = e-151
Identities = 268/363 (73%), Positives = 302/363 (82%), Gaps = 2/363 (0%)
Frame = +2
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNH--HASETAKQGGQTVKPQPIEVPNIS 58
M CF C+ DD A+ G P++ P+GN H HA+ TA + T+ QPI VP+I+
Sbjct: 206 MGCFGFCKGDDSVTVADRG-PFMQSTPTGNPSYHGRHAAVTAPR---TINVQPIAVPSIT 373
Query: 59 EDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSR 118
DELK +TDNFG IGEG+YG+VY LKNG+A IKKLD+S QP+ EFL+QVS+VSR
Sbjct: 374 VDELKPLTDNFGSKCFIGEGAYGKVYQATLKNGRAVVIKKLDSSNQPEHEFLSQVSIVSR 553
Query: 119 LKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAV 178
LKHEN V+L+ Y VDG R L YE+A GSLHDILHGRKGVKGAQPGPVL+WAQRVKIAV
Sbjct: 554 LKHENVVELVNYCVDGPFRALAYEYAPKGSLHDILHGRKGVKGAQPGPVLSWAQRVKIAV 733
Query: 179 GAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLG 238
GAARGLEYLHEKA+ HIIHR IKSSN+L+FDDDVAKIADFDLSNQAPD AARLHSTRVLG
Sbjct: 734 GAARGLEYLHEKAEIHIIHRYIKSSNILLFDDDVAKIADFDLSNQAPDAAARLHSTRVLG 913
Query: 239 TFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSED 298
TFGYHAPEYAMTGQL +KSDVYSFGV+LLELLTGRKPVDHTLPRGQQSLVTWATPKLSED
Sbjct: 914 TFGYHAPEYAMTGQLTSKSDVYSFGVILLELLTGRKPVDHTLPRGQQSLVTWATPKLSED 1093
Query: 299 KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAGE 358
KV+QCVD RL GEYP K+VAKMAAVAALCVQYEA+FRPNMSI+VKALQPLL R + E
Sbjct: 1094KVKQCVDVRLKGEYPSKSVAKMAAVAALCVQYEAEFRPNMSIIVKALQPLLNTRSSHSKE 1273
Query: 359 TAN 361
++N
Sbjct: 1274SSN 1282
>TC216763 UP|Q9LKY3 (Q9LKY3) Pti1 kinase-like protein, complete
Length = 1634
Score = 529 bits (1362), Expect = e-151
Identities = 263/352 (74%), Positives = 296/352 (83%)
Frame = +1
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
M CF C+ DD A+ G P++ P+GN ++H TA +T+ QPI VP+I+ D
Sbjct: 196 MGCFGFCKGDDSVTVADRG-PFMQSTPTGNP-SYHGRHTAVTAPRTINFQPIAVPSITVD 369
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
ELK +TDNFG IGEG+YG+VY LKNG A IKKLD+S QP++EFL+QVS+VSRLK
Sbjct: 370 ELKSLTDNFGSKYFIGEGAYGKVYQATLKNGHAVVIKKLDSSNQPEQEFLSQVSIVSRLK 549
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
HEN V+L+ Y VDG R L YE+A GSLHDILHGRKGVKGAQPGPVL+WAQRVKIAVGA
Sbjct: 550 HENVVELVNYCVDGPFRALAYEYAPKGSLHDILHGRKGVKGAQPGPVLSWAQRVKIAVGA 729
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
ARGLEYLHEKA+ HIIHR IKSSN+L+FDDDVAK+ADFDLSNQAPD AARLHSTRVLGTF
Sbjct: 730 ARGLEYLHEKAEIHIIHRYIKSSNILLFDDDVAKVADFDLSNQAPDAAARLHSTRVLGTF 909
Query: 241 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
GYHAPEYAMTGQL +KSDVYSFGV+LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV
Sbjct: 910 GYHAPEYAMTGQLTSKSDVYSFGVILLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 1089
Query: 301 RQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTAR 352
+QCVD RL GEYP K+VAKMAAVAALCVQYEA+FRPNMSI+VKALQPLL R
Sbjct: 1090KQCVDVRLKGEYPSKSVAKMAAVAALCVQYEAEFRPNMSIIVKALQPLLNTR 1245
>TC226549 UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1627
Score = 493 bits (1269), Expect = e-140
Identities = 258/359 (71%), Positives = 288/359 (79%), Gaps = 5/359 (1%)
Frame = +3
Query: 3 CFNCCEEDDFHKTAESGGPYVVKNPSG-NDGNHHASE-TAKQGGQTVK-PQPIEVPNISE 59
C C E+ + +K+P DGN S+ +A +T K P PIE P +S
Sbjct: 213 CCTCQVEESYPSNENEH----LKSPRNYGDGNQKGSKVSAPVKPETQKAPPPIEAPALSL 380
Query: 60 DELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE--EFLAQVSMVS 117
DELKE TDNFG +LIGEGSYGRVYY L NG+A A+KKLD S +P+ EFL QVSMVS
Sbjct: 381 DELKEKTDNFGSKALIGEGSYGRVYYATLNNGKAVAVKKLDVSSEPESNNEFLTQVSMVS 560
Query: 118 RLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIA 177
RLK+ NFV+L GY V+GN R+L YEFA+ GSLHDILHGRKGV+GAQPGP L W QRV+IA
Sbjct: 561 RLKNGNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIA 740
Query: 178 VGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVL 237
V AARGLEYLHEK P IIHRDI+SSNVLIF+D AKIADF+LSNQAPDMAARLHSTRVL
Sbjct: 741 VDAARGLEYLHEKVQPPIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVL 920
Query: 238 GTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE 297
GTFGYHAPEYAMTGQL KSDVYSFGVVLLELLTGRKPVDHT+PRGQQSLVTWATP+LSE
Sbjct: 921 GTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSE 1100
Query: 298 DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPA 356
DKV+QCVD +L GEYPPK VAK+ AVAALCVQYEA+FRPNMSIVVKALQPLL + P PA
Sbjct: 1101DKVKQCVDPKLKGEYPPKGVAKLGAVAALCVQYEAEFRPNMSIVVKALQPLLKS-PAPA 1274
>TC221155 similar to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein kinase-like
protein (At3g62220), partial (51%)
Length = 724
Score = 345 bits (886), Expect = 1e-95
Identities = 166/196 (84%), Positives = 179/196 (90%)
Frame = +3
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCF CCEEDD+ KTAESGG +VVKN +GNDGN ASETAKQG Q VK QPIEVP + D
Sbjct: 135 MSCFGCCEEDDYQKTAESGGQHVVKNSTGNDGNSRASETAKQGTQAVKIQPIEVPELQVD 314
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
ELKE+TD FG+ SLIGEGSYGRVYYGVLK+GQAAAIKKLDASKQPD+EFLAQVSMVSRLK
Sbjct: 315 ELKEITDGFGESSLIGEGSYGRVYYGVLKSGQAAAIKKLDASKQPDDEFLAQVSMVSRLK 494
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
H+NFVQLLGY +DGNSR+L YEFASNGSLHDILHGRKGVKGAQPGPVLTW QRVKIAVGA
Sbjct: 495 HDNFVQLLGYCIDGNSRVLAYEFASNGSLHDILHGRKGVKGAQPGPVLTWTQRVKIAVGA 674
Query: 181 ARGLEYLHEKADPHII 196
A+GLEYLHE+ADPHII
Sbjct: 675 AKGLEYLHERADPHII 722
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 261 bits (668), Expect = 3e-70
Identities = 146/289 (50%), Positives = 195/289 (66%), Gaps = 3/289 (1%)
Frame = +1
Query: 71 QDSLIGEGSYGRVYYGVLKNGQAAAIKKLD-ASKQPDEEFLAQVSMVSRLKHENFVQLLG 129
+ ++IG G +G VY GVL +G+ AIK +D A KQ +EEF +V ++SRL + LLG
Sbjct: 4 KSNVIGHGGFGLVYRGVLNDGRKVAIKFMDQAGKQGEEEFKVEVELLSRLHSPYLLALLG 183
Query: 130 YSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPV-LTWAQRVKIAVGAARGLEYLH 188
Y D N ++LVYEF +NG L + L+ V + PV L W R++IA+ AA+GLEYLH
Sbjct: 184 YCSDSNHKLLVYEFMANGGLQEHLYP---VSNSIITPVKLDWETRLRIALEAAKGLEYLH 354
Query: 189 EKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYA 248
E P +IHRD KSSN+L+ AK++DF L+ PD A STRVLGT GY APEYA
Sbjct: 355 EHVSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYA 534
Query: 249 MTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE-DKVRQCVDTR 307
+TG L KSDVYS+GVVLLELLTGR PVD P G+ LV+WA P L++ +KV + +D
Sbjct: 535 LTGHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPS 714
Query: 308 LGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPA 356
L G+Y K V ++AA+AA+CVQ EAD+RP M+ VV++L PL+ + P+
Sbjct: 715 LEGQYSMKEVVQVAAIAAMCVQPEADYRPLMADVVQSLVPLVKTQRSPS 861
>TC226480 similar to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein kinase-like
protein (At3g62220), partial (37%)
Length = 739
Score = 258 bits (660), Expect = 2e-69
Identities = 128/135 (94%), Positives = 128/135 (94%)
Frame = +2
Query: 227 MAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQS 286
M A H TRVL FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQS
Sbjct: 2 MXAXXHCTRVLXXFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQS 181
Query: 287 LVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
LVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ
Sbjct: 182 LVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 361
Query: 347 PLLTARPGPAGETAN 361
PLLTARPGPAGET N
Sbjct: 362 PLLTARPGPAGETPN 406
>TC229211 similar to GB|AAR99874.1|41323411|AY518291 strubbelig receptor
family 6 {Arabidopsis thaliana;} , partial (42%)
Length = 1195
Score = 246 bits (628), Expect = 1e-65
Identities = 137/303 (45%), Positives = 194/303 (63%), Gaps = 4/303 (1%)
Frame = +3
Query: 51 PIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE--- 107
P V + S EL+ T +F D L+GEGS+GRVY+ +GQ A+KK+D+S P++
Sbjct: 3 PANVKSYSIAELQIATGSFSVDHLVGEGSFGRVYHAQFDDGQVLAVKKIDSSILPNDLTD 182
Query: 108 EFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPV 167
+F+ +S +S L H N +L+GY + +LVYEF NGSLHD LH +
Sbjct: 183 DFIQIISNISNLHHPNVTELVGYCSEYGQHLLVYEFHKNGSLHDFLH-----LSDEYSKP 347
Query: 168 LTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDM 227
L W RVKIA+G AR LEYLHE + P ++H++IKS+N+L+ + ++D L++ P+
Sbjct: 348 LIWNSRVKIALGTARALEYLHEVSSPSVVHKNIKSANILLDTELNPHLSDSGLASYIPNA 527
Query: 228 AARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSL 287
L+ +G+ GY APE A++GQ KSDVYSFGVV+LELL+GR P D + PR +QSL
Sbjct: 528 DQILNHN--VGS-GYDAPEVALSGQYTLKSDVYSFGVVMLELLSGRNPFDSSRPRSEQSL 698
Query: 288 VTWATPKLSE-DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
V WATP+L + D + + VD + G YP K++++ A V ALCVQ E +FRP MS VV+AL
Sbjct: 699 VRWATPQLHDIDALAKMVDPAMKGLYPVKSLSRFADVIALCVQPEPEFRPPMSEVVQALV 878
Query: 347 PLL 349
L+
Sbjct: 879 RLV 887
>TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (86%)
Length = 1730
Score = 144 bits (364), Expect(2) = 1e-65
Identities = 79/162 (48%), Positives = 104/162 (63%), Gaps = 1/162 (0%)
Frame = +3
Query: 197 HRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAK 256
+RD KSSN+L+ + K++DF L+ P STRV+GT+GY APEYAMTGQL K
Sbjct: 657 NRDCKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVK 836
Query: 257 SDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE-DKVRQCVDTRLGGEYPPK 315
SDVYSFGVV LEL+TGRK +D T P+G+Q+LVTWA P ++ K + D RL G +P +
Sbjct: 837 SDVYSFGVVFLELITGRKAIDSTQPQGEQNLVTWARPLFNDRRKFSKLADPRLQGRFPMR 1016
Query: 316 AVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAG 357
+ + AVA++C+Q A RP + VV AL L P G
Sbjct: 1017GLYQALAVASMCIQESAATRPLIGDVVTALSYLANQAYDPNG 1142
Score = 123 bits (309), Expect(2) = 1e-65
Identities = 80/201 (39%), Positives = 110/201 (53%), Gaps = 9/201 (4%)
Frame = +2
Query: 2 SCFNCCEEDDF-----HKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVP- 55
SCF+ ++D H+ + + + PSG D S + P +++
Sbjct: 59 SCFDSSSKEDHNLRPQHQPNQPLPSQISRLPSGADKLRSRSNGGSKRELQQPPPTVQIAA 238
Query: 56 -NISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKN-GQAAAIKKLDASK-QPDEEFLAQ 112
+ EL T NF +S +GEG +GRVY G L+ Q A+K+LD + Q + EFL +
Sbjct: 239 QTFTFRELAAATKNFRPESFVGEGGFGRVYKGRLETTAQIVAVKQLDKNGLQGNREFLVE 418
Query: 113 VSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQ 172
V M+S L H N V L+GY DG+ R+LVYEF GSL D LH K +P L W
Sbjct: 419 VLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDK--EP---LDWNT 583
Query: 173 RVKIAVGAARGLEYLHEKADP 193
R+KIAVGAA+GLEYLH+KA+P
Sbjct: 584 RMKIAVGAAKGLEYLHDKANP 646
>TC205020 similar to GB|AAR99874.1|41323411|AY518291 strubbelig receptor
family 6 {Arabidopsis thaliana;} , partial (50%)
Length = 1841
Score = 237 bits (605), Expect = 5e-63
Identities = 133/303 (43%), Positives = 188/303 (61%), Gaps = 4/303 (1%)
Frame = +2
Query: 51 PIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE--- 107
P V + S +L+ T F + L+GEGS+GRVY +G+ A+KK+D+S P++
Sbjct: 524 PTNVKSYSIADLQIATGTFSVEQLLGEGSFGRVYRAQFDDGKVLAVKKIDSSVLPNDMSD 703
Query: 108 EFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPV 167
+F+ VS +S+L N +L+GY + +LVYEF NGSLHD LH +
Sbjct: 704 DFVELVSNISQLHDPNVTELVGYCSEHGQHLLVYEFHKNGSLHDFLH-----LPDEYSKP 868
Query: 168 LTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDM 227
L W RVKIA+G AR LEYLHE P ++H++IKS+N+L+ D ++D L++ P+
Sbjct: 869 LIWNSRVKIALGIARALEYLHEVCSPSVVHKNIKSANILLDTDFNPHLSDSGLASYIPNA 1048
Query: 228 AARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSL 287
L++ GY APE ++G KSDVYSFGVV+LELL+GRKP D + PR +Q+L
Sbjct: 1049NQVLNNN---AGSGYEAPEVGLSGHYTLKSDVYSFGVVMLELLSGRKPFDSSRPRSEQAL 1219
Query: 288 VTWATPKLSE-DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
V WATP+L + D + + VD L G YP K++++ A V ALCVQ E +FRP MS VV+AL
Sbjct: 1220VRWATPQLHDIDALAKMVDPALEGLYPVKSLSRFADVIALCVQPEPEFRPPMSEVVQALV 1399
Query: 347 PLL 349
L+
Sbjct: 1400RLV 1408
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 233 bits (594), Expect = 1e-61
Identities = 134/305 (43%), Positives = 191/305 (61%), Gaps = 12/305 (3%)
Frame = +1
Query: 60 DELKEVTDNFGQDSLIGEGSYGRVYYGVLK----------NGQAAAIKKLDASK-QPDEE 108
+ELK T NF DSL+GEG +G VY G + +G A+KKL Q +E
Sbjct: 448 NELKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGLQGHKE 627
Query: 109 FLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVL 168
+L +V + +L H+N V+L+GY DG +R+LVYEF S GSL + L R G QP L
Sbjct: 628 WLTEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFRR----GPQP---L 786
Query: 169 TWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMA 228
+W+ R+K+A+GAARGL +LH A +I+RD K+SN+L+ + AK++DF L+ P
Sbjct: 787 SWSVRMKVAIGAARGLSFLHN-AKSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGD 963
Query: 229 ARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLV 288
ST+V+GT GY APEY TG+L AKSDVYSFGVVLLELL+GR+ VD + +Q+LV
Sbjct: 964 RTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLV 1143
Query: 289 TWATPKLSED-KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQP 347
WA P L + ++ + +DT+LGG+YP K A +A C+ EA RP ++ V++ L+
Sbjct: 1144EWAKPYLGDKRRLFRIMDTKLGGQYPQKGAYMAATLALKCLNREAKGRPPITEVLQTLEQ 1323
Query: 348 LLTAR 352
+ ++
Sbjct: 1324IAASK 1338
>TC226483 homologue to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein
kinase-like protein (At3g62220), partial (39%)
Length = 595
Score = 231 bits (589), Expect = 4e-61
Identities = 113/118 (95%), Positives = 116/118 (97%)
Frame = +2
Query: 218 FDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVD 277
FDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVD
Sbjct: 2 FDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVD 181
Query: 278 HTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFR 335
HTLPRGQQSLVTWATP+LSEDKVRQCVD RLGGEYPPKAVAKMAAVAALCVQYE+ F+
Sbjct: 182 HTLPRGQQSLVTWATPRLSEDKVRQCVDARLGGEYPPKAVAKMAAVAALCVQYES*FQ 355
Score = 58.2 bits (139), Expect = 6e-09
Identities = 29/48 (60%), Positives = 33/48 (68%)
Frame = +3
Query: 314 PKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAGETAN 361
PK + + + +ADFRPNMSIVVKALQPLL AR GPAGET N
Sbjct: 291 PKLLLRWPLLLPCACNTKADFRPNMSIVVKALQPLLNARHGPAGETPN 434
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 230 bits (587), Expect = 7e-61
Identities = 128/298 (42%), Positives = 187/298 (61%), Gaps = 9/298 (3%)
Frame = +2
Query: 60 DELKEVTDNFGQDSLIGEGSYGRVYYGV----LKNG---QAAAIKKLDASK-QPDEEFLA 111
+EL+E T++F +++GEG +G VY G L++G Q A+K+LD Q E+LA
Sbjct: 44 EELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLDLDGLQGHREWLA 223
Query: 112 QVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWA 171
++ + +L+H + V+L+GY + R+L+YE+ GSL + L R + W+
Sbjct: 224 EIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYSA-------AMPWS 382
Query: 172 QRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARL 231
R+KIA+GAA+GL +LHE AD +I+RD K+SN+L+ D AK++DF L+ P+
Sbjct: 383 TRMKIALGAAKGLAFLHE-ADKPVIYRDFKASNILLDSDFTAKLSDFGLAKDGPEGEDTH 559
Query: 232 HSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWA 291
+TR++GT GY APEY MTG L KSDVYS+GVVLLELLTGR+ VD + +SLV WA
Sbjct: 560 VTTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGKSLVEWA 739
Query: 292 TPKL-SEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
P L + KV +D RL G++P K K+A +A C+ + + RP MS V+K L+PL
Sbjct: 740 RPLLRDQKKVYNIIDRRLEGQFPMKGAMKVAMLAFKCLSHHPNARPTMSDVIKVLEPL 913
>TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {Arabidopsis
thaliana;} , partial (42%)
Length = 1533
Score = 229 bits (584), Expect = 1e-60
Identities = 119/286 (41%), Positives = 175/286 (60%)
Frame = +2
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
EL T NF D+LIG G VY G L +G+ A+K L S+ +EF+ ++ +++ L+
Sbjct: 434 ELLSATSNFASDNLIGRGGCSYVYRGCLPDGKELAVKILKPSENVIKEFVQEIEIITTLR 613
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
H+N + + G+ ++GN +LVY+F S GSL + LHG K A W +R K+AVG
Sbjct: 614 HKNIISISGFCLEGNHLLLVYDFLSRGSLEENLHGNKVDCSA-----FGWQERYKVAVGV 778
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
A L+YLH +IHRD+KSSN+L+ DD +++DF L++ ++ + T V GTF
Sbjct: 779 AEALDYLHNGCAQAVIHRDVKSSNILLADDFEPQLSDFGLASWGSS-SSHITCTDVAGTF 955
Query: 241 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
GY APEY M G++ K DVY+FGVVLLELL+ RKP+++ P+GQ+SLV WATP L K
Sbjct: 956 GYLAPEYFMHGRVTDKIDVYAFGVVLLELLSNRKPINNESPKGQESLVMWATPILEGGKF 1135
Query: 301 RQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
Q +D LG EY + +M A LC++ RP +++ + L+
Sbjct: 1136SQLLDPSLGSEYNTCQIKRMILAATLCIRRIPRLRPQINLALLDLE 1273
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 229 bits (584), Expect = 1e-60
Identities = 128/288 (44%), Positives = 178/288 (61%), Gaps = 2/288 (0%)
Frame = +3
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLD-ASKQPDEEFLAQVSMVSRL 119
EL+ T F Q + + EG +G V+ GVL +GQ A+K+ AS Q D+EF ++V ++S
Sbjct: 18 ELQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLASTQGDKEFCSEVEVLSCA 197
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
+H N V L+G+ VD R+LVYE+ NGSL ++ RK VL W+ R KIAVG
Sbjct: 198 QHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRKQ-------NVLEWSARQKIAVG 356
Query: 180 AARGLEYLHEKADPH-IIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLG 238
AARGL YLHE+ I+HRD++ +N+L+ D A + DF L+ PD + TRV+G
Sbjct: 357 AARGLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQPDGDMGVE-TRVIG 533
Query: 239 TFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSED 298
TFGY APEYA +GQ+ K+DVYSFG+VLLEL+TGRK VD P+GQQ L WA P L +
Sbjct: 534 TFGYLAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEWARPLLEKQ 713
Query: 299 KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
+ +D L Y + V +M ++LC+ + RP MS V++ L+
Sbjct: 714 ATYKLIDPSLRNCYVDQEVYRMLKCSSLCIGRDPHLRPRMSQVLRMLE 857
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 220 bits (560), Expect = 9e-58
Identities = 128/342 (37%), Positives = 199/342 (57%), Gaps = 5/342 (1%)
Frame = +1
Query: 13 HKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQD 72
H+ + G + S G ++ G ++ P S L++VT+NF ++
Sbjct: 1462 HENGKGGFKLDAVHVSNGYGGVPVELQSQSSGDRSDLHALDGPTFSIQVLQQVTNNFSEE 1641
Query: 73 SLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE---EFLAQVSMVSRLKHENFVQLLG 129
+++G G +G VY G L +G A+K++++ ++ EF A+++++S+++H + V LLG
Sbjct: 1642 NILGRGGFGVVYKGQLHDGTKIAVKRMESVAMGNKGLKEFEAEIAVLSKVRHRHLVALLG 1821
Query: 130 YSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHE 189
Y ++G R+LVYE+ G+L L + +G P LTW QRV IA+ ARG+EYLH
Sbjct: 1822 YCINGIERLLVYEYMPQGTLTQHLFEWQE-QGYVP---LTWKQRVVIALDVARGVEYLHS 1989
Query: 190 KADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAM 249
A IHRD+K SN+L+ DD AK+ADF L APD + TR+ GTFGY APEYA
Sbjct: 1990 LAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE-TRLAGTFGYLAPEYAA 2166
Query: 250 TGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL-SEDKVRQCVDTRL 308
TG++ K D+Y+FG+VL+EL+TGRK +D T+P + LVTW L +++ + + +D L
Sbjct: 2167 TGRVTTKVDIYAFGIVLMELITGRKALDDTVPDERSHLVTWFRRVLINKENIPKAIDQTL 2346
Query: 309 G-GEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 349
E +++ K+A +A C E RP+M V L PL+
Sbjct: 2347 NPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLVPLV 2472
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 219 bits (559), Expect = 1e-57
Identities = 128/305 (41%), Positives = 184/305 (59%), Gaps = 10/305 (3%)
Frame = +1
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLK--------NGQAAAIKKLDA-SKQPDEEFLA 111
ELK T NF D+++GEG +G+V+ G L+ +G A+KKL++ S Q EE+ +
Sbjct: 124 ELKAATRNFKADTVLGEGGFGKVFKGWLEEKATSKGGSGTVIAVKKLNSESLQGLEEWQS 303
Query: 112 QVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWA 171
+V+ + RL H N V+LLGY ++ + +LVYEF GSL + L GR QP P W
Sbjct: 304 EVNFLGRLSHTNLVKLLGYCLEESELLLVYEFMQKGSLENHLFGRGSA--VQPLP---WD 468
Query: 172 QRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARL 231
R+KIA+GAARGL +LH +I+RD K+SN+L+ AKI+DF L+ P +
Sbjct: 469 IRLKIAIGAARGLAFLH--TSEKVIYRDFKASNILLDGSYNAKISDFGLAKLGPSASQSH 642
Query: 232 HSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWA 291
+TRV+GT GY APEY TG L KSDVY FGVVL+E+LTG + +D P GQ L W
Sbjct: 643 VTTRVMGTHGYAAPEYVATGHLYVKSDVYGFGVVLVEILTGLRALDSNRPSGQHKLTEWV 822
Query: 292 TPKLSE-DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLT 350
P L + K++ +D+RL G++P KA ++A ++ C+ E RP+M V++ L+ +
Sbjct: 823 KPYLHDRRKLKGIMDSRLEGKFPSKAAFRIAQLSMKCLASEPKHRPSMKDVLENLERIQA 1002
Query: 351 ARPGP 355
A P
Sbjct: 1003ANEKP 1017
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 217 bits (553), Expect = 6e-57
Identities = 134/339 (39%), Positives = 194/339 (56%), Gaps = 21/339 (6%)
Frame = +1
Query: 31 DGNHHASETAKQGGQTVKPQP------IEVPNISE---DELKEVTDNFGQDSLIGEGSYG 81
DGN S K ++ P ++ N+ ELK T NF DS++GEG +G
Sbjct: 139 DGNDLGSTNDKVSANSIPQTPRSEGEILQSSNLKSFTLSELKTATRNFRPDSVLGEGGFG 318
Query: 82 RVYYGVLKN----------GQAAAIKKLDASKQPDE-EFLAQVSMVSRLKHENFVQLLGY 130
V+ G + G A+K+L+ P E+LA+V+ + + H + V+L+G+
Sbjct: 319 SVFKGWIDENSLTATKPGTGIVIAVKRLNQDGIPGHREWLAEVNYLGQHFHSHLVRLIGF 498
Query: 131 SVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEK 190
++ R+LVYEF GS + L R QP L+W+ R+K+A+ AA+GL +LH
Sbjct: 499 CLEDEHRLLVYEFMPRGSWENHLFRRGSY--FQP---LSWSLRLKVALDAAKGLAFLHS- 660
Query: 191 ADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMT 250
A+ +I+RD K+SNVL+ AK++DF L+ P STRV+GT+GY APEY T
Sbjct: 661 AEAKVIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPTGDKSHVSTRVMGTYGYAAPEYLAT 840
Query: 251 GQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL-SEDKVRQCVDTRLG 309
G L AKSDVYSFGVVLLE+L+G++ VD P GQ +LV WA P + ++ K+ + +DTRL
Sbjct: 841 GHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHNLVEWAKPFMANKRKIFRVLDTRLQ 1020
Query: 310 GEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
G+Y K+A +A C+ E+ FRPNM VV L+ L
Sbjct: 1021GQYSTDDAYKLATLALRCLSIESKFRPNMDQVVTTLEQL 1137
>AW707099 weakly similar to GP|3668069|gb| Pto kinase interactor 1
{Lycopersicon esculentum}, partial (33%)
Length = 463
Score = 216 bits (550), Expect = 1e-56
Identities = 102/153 (66%), Positives = 128/153 (82%)
Frame = +2
Query: 203 SNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSF 262
SNVLIF+DDVAKIADF+LSNQAP++ RLHSTRVLGT GYH PEYA+T QLN KS+VY F
Sbjct: 2 SNVLIFNDDVAKIADFNLSNQAPNITTRLHSTRVLGTLGYHTPEYAITKQLNTKSNVYRF 181
Query: 263 GVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAA 322
++LLELLT +KP++HTLPRGQQS +T TPKLS++KVRQ ++TRL YPPKA+ KM
Sbjct: 182 NIILLELLTRKKPINHTLPRGQQSAIT*TTPKLSKNKVRQYINTRLKE*YPPKAITKMTT 361
Query: 323 VAALCVQYEADFRPNMSIVVKALQPLLTARPGP 355
+ L +QY+ADF+PN+S++ KA+QP+LT RP P
Sbjct: 362 ITTLYMQYKADFKPNISMIXKAVQPVLTTRPRP 460
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 216 bits (549), Expect = 2e-56
Identities = 130/312 (41%), Positives = 187/312 (59%), Gaps = 13/312 (4%)
Frame = +3
Query: 50 QPIEVPNISEDELKEVTDNFGQDSLIG-EGSYGRVYYGVLKN----------GQAAAIKK 98
Q + N S EL T NF +DS++G EG +G V+ G + N G A+K+
Sbjct: 285 QSSNLKNFSLTELTAATRNFRKDSVLGGEGDFGSVFKGWIDNQSLAAAKPGTGVVVAVKR 464
Query: 99 LDA-SKQPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRK 157
L S Q ++ LA+V+ + +L H + V+L+GY + R+LVYEF GSL + L
Sbjct: 465 LSLDSFQGHKDRLAEVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLF--- 635
Query: 158 GVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIAD 217
++G+ P L+W R+K+A+GAA+GL +LH A+ +I+RD K+SNVL+ + AK+AD
Sbjct: 636 -MRGSYFQP-LSWGLRLKVALGAAKGLAFLHS-AETKVIYRDFKTSNVLLDSNYNAKLAD 806
Query: 218 FDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVD 277
L+ P STRV+GT+GY APEY TG L+AKSDV+SFGVVLLE+L+GR+ VD
Sbjct: 807 LGLAKDGPTREKSHASTRVMGTYGYAAPEYLATGNLSAKSDVFSFGVVLLEMLSGRRAVD 986
Query: 278 HTLPRGQQSLVTWATPKLS-EDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRP 336
P GQ +LV WA P LS + K+ + +D RL G+Y K+A ++ C+ E+ RP
Sbjct: 987 KNRPSGQHNLVEWAKPYLSNKRKLLRVLDNRLEGQYELDEACKVATLSLRCLAIESKLRP 1166
Query: 337 NMSIVVKALQPL 348
M V L+ L
Sbjct: 1167TMDEVATDLEQL 1202
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,345,808
Number of Sequences: 63676
Number of extensions: 180452
Number of successful extensions: 2603
Number of sequences better than 10.0: 965
Number of HSP's better than 10.0 without gapping: 1860
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1883
length of query: 361
length of database: 12,639,632
effective HSP length: 98
effective length of query: 263
effective length of database: 6,399,384
effective search space: 1683037992
effective search space used: 1683037992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0158.11


