

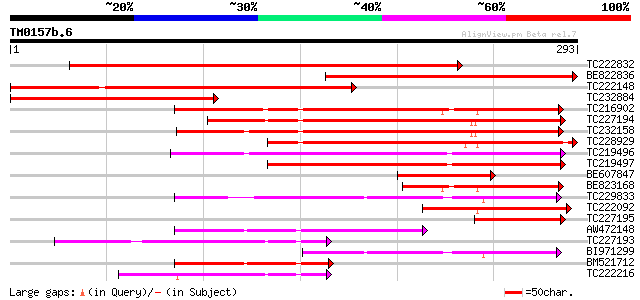
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0157b.6
(293 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222832 similar to UP|Q75ZI4 (Q75ZI4) Prolyl 4-hydroxylase, par... 345 2e-95
BE822836 205 2e-53
TC222148 weakly similar to UP|Q75ZI4 (Q75ZI4) Prolyl 4-hydroxyla... 185 2e-47
TC232884 similar to UP|Q75ZI4 (Q75ZI4) Prolyl 4-hydroxylase, par... 181 4e-46
TC216902 similar to UP|Q8LAN3 (Q8LAN3) Prolyl 4-hydroxylase alph... 166 9e-42
TC227194 similar to UP|Q8T5S8 (Q8T5S8) Prolyl 4-hydroxylase alph... 158 2e-39
TC232158 153 1e-37
TC228929 147 4e-36
TC219496 144 4e-35
TC219497 125 2e-29
BE607847 94 6e-20
BE823168 87 9e-18
TC229833 79 2e-15
TC222092 similar to UP|Q8VZD7 (Q8VZD7) AT3g28480/MFJ20_16, parti... 76 2e-14
TC227195 70 1e-12
AW472148 69 2e-12
TC227193 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alp... 59 3e-09
BI971299 57 1e-08
BM521712 similar to GP|21618073|gb| prolyl 4-hydroxylase alpha s... 55 3e-08
TC222216 similar to UP|Q9LSI6 (Q9LSI6) Prolyl 4-hydroxylase alph... 54 8e-08
>TC222832 similar to UP|Q75ZI4 (Q75ZI4) Prolyl 4-hydroxylase, partial (57%)
Length = 619
Score = 345 bits (884), Expect = 2e-95
Identities = 170/203 (83%), Positives = 184/203 (89%)
Frame = +1
Query: 32 AGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSIPFQVLSWKPR 91
A F TLFSHSQ DG GLRPR RLL+S+ +AEYNLM GE GDDSITSIPFQVLSW+PR
Sbjct: 7 AEFGTRTLFSHSQGDGDGLRPRQRLLDSTNEAEYNLMPVGELGDDSITSIPFQVLSWRPR 186
Query: 92 ALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGT 151
A+YFPNFATAEQC+SI+ VAK GLKPS LALR+GETE NTKGIRTSSGVFVS+SEDKT T
Sbjct: 187 AVYFPNFATAEQCESIIDVAKDGLKPSTLALRQGETEDNTKGIRTSSGVFVSASEDKTRT 366
Query: 152 LAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYL 211
L +IEEKIARATMIPRSHGEAFNILRYEV+QRYN HYD+FNPAEYGPQKSQRMASFLLYL
Sbjct: 367 LDVIEEKIARATMIPRSHGEAFNILRYEVNQRYNSHYDAFNPAEYGPQKSQRMASFLLYL 546
Query: 212 TDVQEGGETMFPFENGLNMDVSY 234
TDV+EGGETMFPFENGLNMD +Y
Sbjct: 547 TDVEEGGETMFPFENGLNMDGNY 615
>BE822836
Length = 610
Score = 205 bits (521), Expect = 2e-53
Identities = 93/130 (71%), Positives = 109/130 (83%)
Frame = -2
Query: 164 MIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFP 223
MIP +HG FNIL+YEV Q+Y+ HYD+FNP EY +SQR+ASFLLYL++V+ GGETMFP
Sbjct: 609 MIPXTHGXIFNILKYEVXQKYDSHYDAFNPDEYXXVESQRIASFLLYLSNVEAGGETMFP 430
Query: 224 FENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVAT 283
+E GLN+D Y Y+ CIGL+V+PRQGDGLLFYSL PNG ID TSLHGSCPVIKGEKWVAT
Sbjct: 429 YEGGLNIDRGYDYQKCIGLKVKPRQGDGLLFYSLLPNGKIDKTSLHGSCPVIKGEKWVAT 250
Query: 284 KWIRDHDQDY 293
KWI D +Q Y
Sbjct: 249 KWIDDREQHY 220
>TC222148 weakly similar to UP|Q75ZI4 (Q75ZI4) Prolyl 4-hydroxylase, partial
(40%)
Length = 619
Score = 185 bits (469), Expect = 2e-47
Identities = 90/179 (50%), Positives = 124/179 (68%)
Frame = +1
Query: 1 MKAKTVKGNLSSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESS 60
++ + +KG + S KL P +F++C FF GFF S L DD + PR R+L+ S
Sbjct: 91 LQREMMKGKVKSSKLKLGVPTLFILCALFFFVGFFVSPLLFQDLDD---VGPRSRILQES 261
Query: 61 EKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSAL 120
K EY + GE G+ + SIP Q+LSW+PRA++FPNF + E C+ I+ +AK L+PS L
Sbjct: 262 VKKEYEPLEHGESGEPFVDSIPSQILSWRPRAVFFPNFTSVEVCQQIIEMAKPKLEPSKL 441
Query: 121 ALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYE 179
ALR+GET ++TK RTSSG F+S+SEDK+G L ++E KIA+ TMIPR+HGE FNIL+YE
Sbjct: 442 ALRKGETAESTKDTRTSSGTFISASEDKSGILDLVERKIAKVTMIPRTHGEIFNILKYE 618
>TC232884 similar to UP|Q75ZI4 (Q75ZI4) Prolyl 4-hydroxylase, partial (10%)
Length = 426
Score = 181 bits (458), Expect = 4e-46
Identities = 85/108 (78%), Positives = 95/108 (87%)
Frame = +2
Query: 1 MKAKTVKGNLSSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESS 60
MKAKTVKGN S R+NKL+FPY+FL+CIFFFLAGFFG TLFSHSQ DG GLRPR RLLE
Sbjct: 101 MKAKTVKGNWSLRTNKLSFPYVFLVCIFFFLAGFFGFTLFSHSQGDGDGLRPRQRLLEPG 280
Query: 61 EKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIV 108
+AEYNLM + GDDSITSIPFQVLSW+PRALYFPNFATAEQC++I+
Sbjct: 281 NEAEYNLMPVRDLGDDSITSIPFQVLSWRPRALYFPNFATAEQCENII 424
>TC216902 similar to UP|Q8LAN3 (Q8LAN3) Prolyl 4-hydroxylase alpha
subunit-like protein, partial (89%)
Length = 1381
Score = 166 bits (421), Expect = 9e-42
Identities = 93/210 (44%), Positives = 132/210 (62%), Gaps = 9/210 (4%)
Frame = +3
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALR-EGETEQNTKGIRTSSGVFVSS 144
+SWKPRA + F T +C ++ +AK+ LK SA+A GE++ + +RTSSG+F+S
Sbjct: 300 ISWKPRAFVYEGFLTDLECDHLISLAKSELKRSAVADNLSGESQLSD--VRTSSGMFISK 473
Query: 145 SEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRM 204
++D +A IE+KI+ T +P+ +GE +LRYE Q+Y+PHYD F + R+
Sbjct: 474 NKDPI--VAGIEDKISSWTFLPKENGEDIQVLRYEHGQKYDPHYDYFTDKVNIARGGHRI 647
Query: 205 ASFLLYLTDVQEGGETMF------PFENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFYS 256
A+ L+YLTDV +GGET+F P G + S +C G+ V+PR+GD LLF+S
Sbjct: 648 ATVLMYLTDVAKGGETVFXSAEEPPRRRG--AETSSDLSECAKKGIAVKPRRGDALLFFS 821
Query: 257 LFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
L N T D +SLH CPVI+GEKW ATKWI
Sbjct: 822 LHTNATPDTSSLHAGCPVIEGEKWSATKWI 911
>TC227194 similar to UP|Q8T5S8 (Q8T5S8) Prolyl 4-hydroxylase alpha-related
protein PH4[alpha]PV (CG31015-PA), partial (4%)
Length = 965
Score = 158 bits (400), Expect = 2e-39
Identities = 82/189 (43%), Positives = 120/189 (63%), Gaps = 4/189 (2%)
Frame = +1
Query: 103 QCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARA 162
+C+ ++ +AK + S + E ++++ +RTSSG F++ DK + IE++IA
Sbjct: 1 ECEYLIDIAKPSMHKSTVVDSETGKSKDSR-VRTSSGTFLARGRDKI--VRDIEKRIAHY 171
Query: 163 TMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMF 222
+ IP HGE +L YEV Q+Y PHYD F QR+A+ L+YLTDV+EGGET+F
Sbjct: 172 SFIPVEHGEGLQVLHYEVGQKYEPHYDYFLDDFNTKNGGQRIATVLMYLTDVEEGGETVF 351
Query: 223 PFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGE 278
P G V + E +C GL ++P++GD LLF+S+ P+ T+DP+SLHG CPVIKG
Sbjct: 352 PAAKGNFSSVPWWNELSECGKKGLSIKPKRGDALLFWSMKPDATLDPSSLHGGCPVIKGN 531
Query: 279 KWVATKWIR 287
KW +TKW+R
Sbjct: 532 KWSSTKWMR 558
>TC232158
Length = 904
Score = 153 bits (386), Expect = 1e-37
Identities = 81/205 (39%), Positives = 125/205 (60%), Gaps = 5/205 (2%)
Frame = +2
Query: 87 SWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNT-KGIRTSSGVFVSSS 145
SW+P A + NF T E+C+ ++ +A ++ S +A + ++ Q+ +R S+G F+
Sbjct: 215 SWQPTAFLYHNFLTKEECEYLINIATPHMQKSTVA--DNQSGQSVVHDVRKSTGAFLDRG 388
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D+ + IE++IA T IP +GE ++ YEV Q Y+PHYD F QR+A
Sbjct: 389 QDEI--VRNIEKRIADVTFIPIENGEPIYVIHYEVGQYYDPHYDYFIDDFNIENGGQRIA 562
Query: 206 SFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFPNG 261
+ L+YL++V+EGGETMFP V + E +C +GL ++P+ GD LLF+S+ PN
Sbjct: 563 TMLMYLSNVEEGGETMFPRAKANFSSVPWWNELSNCGKMGLSIKPKMGDALLFWSMKPNA 742
Query: 262 TIDPTSLHGSCPVIKGEKWVATKWI 286
T+D +LH +CPVIKG KW TKW+
Sbjct: 743 TLDALTLHSACPVIKGNKWSCTKWM 817
>TC228929
Length = 979
Score = 147 bits (372), Expect = 4e-36
Identities = 78/164 (47%), Positives = 104/164 (62%), Gaps = 4/164 (2%)
Frame = +3
Query: 134 IRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNP 193
+RTSSG+F+ DK + IE++IA T IP +GE IL YEV Q+Y PHYD F
Sbjct: 18 VRTSSGMFLKRGRDKI--IQNIEKRIADFTFIPEENGEGLQILHYEVGQKYEPHYDYFLD 191
Query: 194 AEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSY--RYEDCI--GLRVRPRQG 249
QR+A+ L+YL+DV+EGGET+FP N V + C GL V+P+ G
Sbjct: 192 EFNTKNGGQRIATVLMYLSDVEEGGETVFPAANANFSSVPWWNDLSQCARKGLSVKPKMG 371
Query: 250 DGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D LLF+S+ P+ T+DP+SLHG CPVIKG KW +TKW+ H ++Y
Sbjct: 372 DALLFWSMRPDATLDPSSLHGGCPVIKGNKWSSTKWM--HLREY 497
>TC219496
Length = 910
Score = 144 bits (364), Expect = 4e-35
Identities = 81/205 (39%), Positives = 123/205 (59%), Gaps = 1/205 (0%)
Frame = +1
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKG-IRTSSGVFV 142
+VL+W PR + NF + E+C + +A L S + + +T + K +RTSSG+F+
Sbjct: 139 EVLNWSPRIILLHNFLSMEECDYLRALALPRLHISTVV--DTKTGKGIKSDVRTSSGMFL 312
Query: 143 SSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQ 202
+S E K + IE++I+ + IP +GE +LRYE +Q Y PH+D F+ + Q
Sbjct: 313 NSKERKYPMVQAIEKRISVYSQIPIENGELMQVLRYEKNQYYKPHHDYFSDTFNLKRGGQ 492
Query: 203 RMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGT 262
R+A+ L+YL+D EGGET FP + + S + GL V+P +G+ +LF+S+ +G
Sbjct: 493 RIATMLMYLSDNIEGGETYFPLAG--SGECSCGGKLVKGLSVKPIKGNAVLFWSMGLDGQ 666
Query: 263 IDPTSLHGSCPVIKGEKWVATKWIR 287
DP S+HG C VI GEKW ATKW+R
Sbjct: 667 SDPNSVHGGCEVISGEKWSATKWLR 741
>TC219497
Length = 770
Score = 125 bits (314), Expect = 2e-29
Identities = 66/154 (42%), Positives = 97/154 (62%)
Frame = +3
Query: 134 IRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNP 193
+RTSSG+F++ E K + IE++I+ + IP +GE +LRYE +Q Y PH+D F+
Sbjct: 75 VRTSSGMFLNPQERKYPMVQAIEKRISVYSQIPIENGELMQVLRYEKNQYYKPHHDYFSD 254
Query: 194 AEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLL 253
+ QR+A+ L+YL+D EGGET FP + + S + GL V+P +G+ +L
Sbjct: 255 TFNLKRGGQRIATMLMYLSDNIEGGETYFPLAG--SGECSCGGKLVKGLSVKPIKGNAVL 428
Query: 254 FYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
F+S+ +G DP S+HG C VI GEKW ATKW+R
Sbjct: 429 FWSMGLDGQSDPNSVHGGCEVISGEKWSATKWMR 530
>BE607847
Length = 482
Score = 94.4 bits (233), Expect = 6e-20
Identities = 42/51 (82%), Positives = 48/51 (93%)
Frame = +3
Query: 201 SQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDG 251
+ +MASFLLYLTDV+EGGETMFPFENGLNMD +Y YEDCIGL+V+PRQGDG
Sbjct: 330 TMQMASFLLYLTDVEEGGETMFPFENGLNMDGNYGYEDCIGLKVKPRQGDG 482
>BE823168
Length = 654
Score = 87.0 bits (214), Expect = 9e-18
Identities = 45/91 (49%), Positives = 58/91 (63%), Gaps = 8/91 (8%)
Frame = -1
Query: 204 MASFLLYLTDVQEGGETMF------PFENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFY 255
+A L+YLTDV +GGET+F P G + + +C G+ V+PR+GD LLF+
Sbjct: 654 VAXXLMYLTDVXKGGETVFXDAXXSPRHKG--SETNENLXECAQKGIAVKPRRGDALLFF 481
Query: 256 SLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
SL+PN D SLH CPVI+GEKW ATKWI
Sbjct: 480 SLYPNAIPDTLSLHAGCPVIEGEKWSATKWI 388
>TC229833
Length = 1265
Score = 79.3 bits (194), Expect = 2e-15
Identities = 52/203 (25%), Positives = 99/203 (48%), Gaps = 3/203 (1%)
Frame = +3
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+SW+PR + F + ++C +V +A A ++ + G S +S
Sbjct: 294 ISWQPRVFLYKGFLSDKECDYLVSLAYA-------------VKEKSSGNGGLSEGVETSL 434
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+ + LA IEE+++ +P+ + + ++ Y +Q + D F MA
Sbjct: 435 DMEDDILARIEERLSVWAFLPKEYSKPLQVMHYGPEQN-GRNLDYFTNKTQLELSGPLMA 611
Query: 206 SFLLYLT-DVQEGGETMFPFENGLNMDVSYRYEDCIGLR--VRPRQGDGLLFYSLFPNGT 262
+ +LYL+ DV +GG+ +FP ++ S + C ++P +G+ +LF+SL P+ +
Sbjct: 612 TIILYLSNDVTQGGQILFPE----SVPGSSSWSSCSNSSNILQPVKGNAILFFSLHPSAS 779
Query: 263 IDPTSLHGSCPVIKGEKWVATKW 285
D +S H CPV++G+ W A K+
Sbjct: 780 PDKSSFHARCPVLEGDMWSAIKY 848
>TC222092 similar to UP|Q8VZD7 (Q8VZD7) AT3g28480/MFJ20_16, partial (26%)
Length = 455
Score = 76.3 bits (186), Expect = 2e-14
Identities = 40/80 (50%), Positives = 51/80 (63%), Gaps = 3/80 (3%)
Frame = +2
Query: 214 VQEGGETMFP-FENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFYSLFPNGTIDPTSLHG 270
V++GGET+FP E L + +C G V+PR+GD LLF+ L + + D SLHG
Sbjct: 2 VEKGGETIFPNAEAKLLQPKDESWSECAHKGYAVKPRKGDALLFFILHLDASTDTKSLHG 181
Query: 271 SCPVIKGEKWVATKWIRDHD 290
SCPVI+GEKW ATKWI D
Sbjct: 182 SCPVIEGEKWSATKWIHVSD 241
>TC227195
Length = 594
Score = 70.1 bits (170), Expect = 1e-12
Identities = 28/47 (59%), Positives = 39/47 (82%)
Frame = +2
Query: 241 GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
GL ++P++GD LLF+S+ P+ T+DP+SLHG CPVIKG K +TKW+R
Sbjct: 41 GLSIKPKRGDALLFWSMKPDATLDPSSLHGGCPVIKGNKGSSTKWMR 181
>AW472148
Length = 391
Score = 69.3 bits (168), Expect = 2e-12
Identities = 44/131 (33%), Positives = 69/131 (52%)
Frame = +2
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+SWKPRA + F T +C + +AK+ LK SA+A +E I TSS +F+ ++
Sbjct: 8 VSWKPRAFVYEGFLTESECDHFMSIAKSDLKRSAVA-DNLSSESKVSEITTSSCMFIPNN 184
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
D +A IE KI+ T +P + E IL E +Y+PHYD + + + R +
Sbjct: 185 NDL--IVASIEYKISSWTFLPIENEEDIQILICEHGHKYHPHYDYCANKVHITRCTHRSS 358
Query: 206 SFLLYLTDVQE 216
L+ LT+V +
Sbjct: 359 HILISLTNVTD 391
>TC227193 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alpha
subunit-like protein, partial (41%)
Length = 557
Score = 58.9 bits (141), Expect = 3e-09
Identities = 42/144 (29%), Positives = 71/144 (49%), Gaps = 1/144 (0%)
Frame = +1
Query: 24 LICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSIPF 83
L +F FL + RG P+P L S + T DD + +
Sbjct: 142 LFLVFTFLVLILLALGILSIPSSSRGNLPKPNDLASIARN-----TIHTSDDDDVRGEQW 306
Query: 84 -QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFV 142
+V+SW+PRA + NF T E+C+ ++ +AK + S++ E ++++ +RTSSG F+
Sbjct: 307 VEVVSWEPRAFVYHNFLTKEECEYLIDIAKPNMHKSSVVDSETGKSKDSR-VRTSSGTFL 483
Query: 143 SSSEDKTGTLAIIEEKIARATMIP 166
+ DK + IE++IA + IP
Sbjct: 484 ARGRDK--IVRDIEKRIAHYSFIP 549
>BI971299
Length = 669
Score = 57.0 bits (136), Expect = 1e-08
Identities = 36/137 (26%), Positives = 67/137 (48%), Gaps = 3/137 (2%)
Frame = -1
Query: 152 LAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYL 211
LA IEE+++ + + + ++ Y + + D F MA+ +LYL
Sbjct: 648 LARIEERLSXXXXLXXEYSKPXQVMHYGPEPN-GRNLDYFTXKTQLELSGPLMATIVLYL 472
Query: 212 TDVQ-EGGETMFPFENGLNMDVSYRYEDCIGLR--VRPRQGDGLLFYSLFPNGTIDPTSL 268
++ +GG+ +FP ++ S + C ++P +G+ +LF+SL P+ + D S
Sbjct: 471 SNAATQGGQILFPE----SVPRSSSWSSCSNSSNILQPVKGNAILFFSLHPSASPDKNSF 304
Query: 269 HGSCPVIKGEKWVATKW 285
H CPV++G W A K+
Sbjct: 303 HARCPVLEGNMWSAIKY 253
>BM521712 similar to GP|21618073|gb| prolyl 4-hydroxylase alpha subunit-like
protein {Arabidopsis thaliana}, partial (30%)
Length = 412
Score = 55.5 bits (132), Expect = 3e-08
Identities = 30/82 (36%), Positives = 50/82 (60%)
Frame = +1
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+SWKPRA + F T +C ++ +AK+ LK SA+A E +RTSSG+F+ +
Sbjct: 166 VSWKPRAFVYDVFLTELECDHLISIAKSELKRSAVA-DNLSGESKLSEVRTSSGMFIPKN 342
Query: 146 EDKTGTLAIIEEKIARATMIPR 167
+D +A +E+KI+ T++P+
Sbjct: 343 KDP--IVAGVEDKISSWTLLPK 402
>TC222216 similar to UP|Q9LSI6 (Q9LSI6) Prolyl 4-hydroxylase alpha
subunit-like protein, partial (27%)
Length = 573
Score = 53.9 bits (128), Expect = 8e-08
Identities = 36/112 (32%), Positives = 61/112 (54%), Gaps = 2/112 (1%)
Frame = +1
Query: 57 LESSEKAEYNLMTAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAG 114
L+ KA + + G S+ P +V LSW PRA + F + E+C ++ +AK
Sbjct: 247 LDQDAKATHGSVLRLNRGGSSVKFDPTRVTQLSWSPRAFLYKGFLSEEECDHLITLAKDK 426
Query: 115 LKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIP 166
L+ S +A E ++ +RTSSG+F++ ++D+ +A IE +IA T +P
Sbjct: 427 LEKSMVADNESGKSIMSE-VRTSSGMFLNKAQDE--IVAGIEARIAAWTFLP 573
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,225,604
Number of Sequences: 63676
Number of extensions: 179591
Number of successful extensions: 1176
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 1146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1154
length of query: 293
length of database: 12,639,632
effective HSP length: 97
effective length of query: 196
effective length of database: 6,463,060
effective search space: 1266759760
effective search space used: 1266759760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0157b.6


