

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0154.9
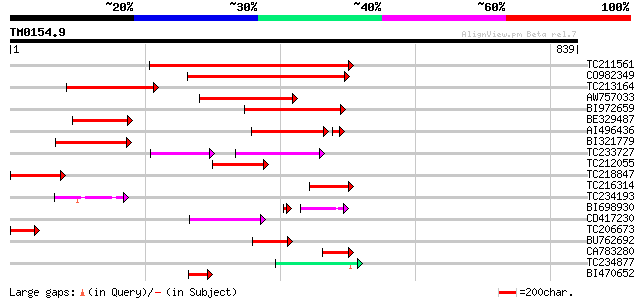
(839 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 321 1e-87
CO982349 228 6e-60
TC213164 154 2e-37
AW757033 139 4e-33
BI972659 133 4e-31
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 129 7e-30
AI496436 111 5e-27
BI321779 118 9e-27
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 59 1e-16
TC212055 84 3e-16
TC218847 77 2e-14
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 74 3e-13
TC234193 70 5e-12
BI698930 63 4e-10
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 61 2e-09
TC206673 59 1e-08
BU762692 57 3e-08
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 52 1e-06
TC234877 45 2e-04
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 45 2e-04
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 321 bits (822), Expect = 1e-87
Identities = 157/301 (52%), Positives = 209/301 (69%)
Frame = +1
Query: 208 LDSVLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGC 267
L S L+ANEVIDEAK + LVFKVDYEK YDSV+WNFL YM+ R+ F KWI WI+ C
Sbjct: 1 LHSALIANEVIDEAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 268 IQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKV 327
++SAS+SVLVNGSPT EF ++ L GDPL PFLF IVAE L+G+MR+A NLYK + V
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLV 360
Query: 328 GREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAED 387
G GV +S+LQ+ADDT+F E + NV A+K +LR FEL+S L++NF KS V +
Sbjct: 361 GANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCFGVFGVTDQ 540
Query: 388 VLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKV 447
AN LHC PF+YLGI +GAN R+ WD ++ K ++LS WK + +SFGG+V
Sbjct: 541 WKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGGRV 720
Query: 448 CLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRME 507
LI+SVL+SIP+++ SFF++P+ V +I + FLWGG +KI+W++W K+C +
Sbjct: 721 TLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPKER 900
Query: 508 G 508
G
Sbjct: 901 G 903
>CO982349
Length = 795
Score = 228 bits (582), Expect = 6e-60
Identities = 109/240 (45%), Positives = 158/240 (65%)
Frame = +3
Query: 263 WIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLY 322
WI+GC+ SASVS+LVN SP +EF ++ L GDPLAP LF IVAE L+G+MR+A +
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRF 248
Query: 323 KGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGV 382
F VG+ VS+LQ+ADDT+F E + N+ +K++LRCFEL SGLK+NF +S+ +
Sbjct: 249 NSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGAI 428
Query: 383 SVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLS 442
+ A L+C +PF YLGI + ANPR + DP++ K +L+ WK++ +S
Sbjct: 429 WKPDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHIS 608
Query: 443 FGGKVCLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVC 502
GG+V LI ++L+++P+++ SFF+ P V I+++FLWGG R+IAWVKW VC
Sbjct: 609 LGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRRIAWVKWETVC 788
>TC213164
Length = 446
Score = 154 bits (389), Expect = 2e-37
Identities = 71/135 (52%), Positives = 99/135 (72%)
Frame = +3
Query: 85 LTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKE 144
L F +E+R +WD KSPGPDG NF FIK FW +LK D++R L++F+ NGV+ K
Sbjct: 18 LVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKR 197
Query: 145 GNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEG 204
N+ F+ LIPKV++P LN+Y PISLIGC+YKIV+K+L+ R KV+ IIDE Q+ F+EG
Sbjct: 198 SNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEG 377
Query: 205 RQLLDSVLVANEVID 219
R L +V++ANE+++
Sbjct: 378 RHTLHNVVIANEIME 422
>AW757033
Length = 441
Score = 139 bits (351), Expect = 4e-33
Identities = 71/145 (48%), Positives = 95/145 (64%)
Frame = -2
Query: 282 TDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFAD 341
T EF + L GDPLAP LF I AE L+G+MR+A N YK F VG+ VS+LQ+AD
Sbjct: 440 TREFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYAD 261
Query: 342 DTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTT 401
DT+F E + NV +K++LR FEL SGLK+NF KS+ +SV + A ++C
Sbjct: 260 DTIFFGEATMENVRVIKTILRGFELASGLKINFAKSRFGAISVPD*WRKEAAEFMNCSLL 81
Query: 402 EIPFVYLGIHVGANPRKKTTWDPLL 426
+PF YLGI +GANPR++ TWDP++
Sbjct: 80 SLPFSYLGIPIGANPRRRETWDPII 6
>BI972659
Length = 453
Score = 133 bits (334), Expect = 4e-31
Identities = 59/149 (39%), Positives = 98/149 (65%)
Frame = +1
Query: 348 EPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVY 407
E S N++ +KSMLR FE++SGL++N+ KS+ V + A LL+C+ +IPF Y
Sbjct: 7 EASWDNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPNWAHDAAQLLNCRQLDIPFHY 186
Query: 408 LGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKL 467
LG+ + + W+PL++K + +LS W +K LS GG+V LI+SVL+++P++ LSFFK+
Sbjct: 187 LGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFKI 366
Query: 468 PKGVASLCNRIEKQFLWGGEEGTRKIAWV 496
P+ + +++ F+WGG + +I+WV
Sbjct: 367 PQRIVDKLVSLQRTFMWGGNQHHNRISWV 453
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 129 bits (323), Expect = 7e-30
Identities = 57/88 (64%), Positives = 69/88 (77%)
Frame = -2
Query: 94 IRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLI 153
I++AVW C KSPGPDG NF FIK FW LK DI+R L +FH NG++PK GN+SFI LI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 154 PKVNNPMGLNDYIPISLIGCMYKIVSKV 181
PKV +P LND+ PISLIGC+YKIV+K+
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKI 112
>AI496436
Length = 414
Score = 111 bits (277), Expect(2) = 5e-27
Identities = 51/113 (45%), Positives = 78/113 (68%)
Frame = +1
Query: 359 SMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRK 418
++LRCFEL SGLK+NF KS+ + ++ A+ L+ +PFVYLGI +GAN R
Sbjct: 1 TILRCFELSSGLKINFAKSQFGTICKSDIWRKEAADYLNSSVLSMPFVYLGIPIGANSRH 180
Query: 419 KTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGV 471
W+P++ K ++L+ WK+K +SFGG+V LI SVL+++P++ LSFF++P V
Sbjct: 181 SDVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKV 339
Score = 29.3 bits (64), Expect(2) = 5e-27
Identities = 11/18 (61%), Positives = 15/18 (83%)
Frame = +2
Query: 478 IEKQFLWGGEEGTRKIAW 495
I+++FLWGG+ RKIAW
Sbjct: 359 IQRRFLWGGDLE*RKIAW 412
>BI321779
Length = 421
Score = 118 bits (296), Expect = 9e-27
Identities = 54/112 (48%), Positives = 72/112 (64%)
Frame = +2
Query: 69 LEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDI 128
L G F S S EDN L + F +EEI+ + DCE KSPGPDGY+ IK FW V+K+++
Sbjct: 35 LNGVEFLSFSNEDNAMLIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIKEEV 214
Query: 129 VRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSK 180
+ ++FH P+ N SFI LI K +NP+ + Y PISL+GC+YKIV K
Sbjct: 215 INFFKEFHKFDFLPRGTNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 58.9 bits (141), Expect(2) = 1e-16
Identities = 32/95 (33%), Positives = 53/95 (55%), Gaps = 1/95 (1%)
Frame = -3
Query: 209 DSVLVANEVIDEAKVKK-RGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGC 267
D V +EVI+ K G L K+D K +D+++ +FL +L + G+ + WI+
Sbjct: 828 DCTCVTSEVINMLDKKVFGGNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRVI 649
Query: 268 IQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLF 302
+ SA +S+ VNG P F ++ + GDPL+P L+
Sbjct: 648 LLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLY 544
Score = 46.6 bits (109), Expect(2) = 1e-16
Identities = 31/131 (23%), Positives = 64/131 (48%)
Frame = -2
Query: 335 SLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYAN 394
S + ADD + + ++ NV + + + ++ G ++ KSK+ S+ L ++
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRLRVISD 272
Query: 395 LLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVL 454
LL ++IPF Y G+ + + + K++ +L+ WK LS G++ L+ SV+
Sbjct: 271 LLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSVI 92
Query: 455 SSIPLFYLSFF 465
+ L+ S +
Sbjct: 91 HGMLLYSFSVY 59
>TC212055
Length = 776
Score = 84.0 bits (206), Expect = 3e-16
Identities = 41/82 (50%), Positives = 58/82 (70%)
Frame = +1
Query: 301 LFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLAMKSM 360
LF IVAE L+G+MR+A +LY VG+ + V++LQ+ADDT+F+ E + NV+ +KSM
Sbjct: 1 LFNIVAEGLTGLMREALDKSLYSSLMVGKNKIPVNILQYADDTIFLGEATMQNVMTIKSM 180
Query: 361 LRCFELMSGLKVNFFKSKLAGV 382
LR FEL SGLK++F KS +
Sbjct: 181 LRVFELASGLKIHFAKSSFGAI 246
>TC218847
Length = 521
Score = 77.4 bits (189), Expect = 2e-14
Identities = 32/82 (39%), Positives = 54/82 (65%)
Frame = -3
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDPKVVKSKMKEFFEARFT 60
QK+R +W+++ D N+ +FH IN+ RTNA+ G+++ G WVE+P VK+ ++ FF+ RF+
Sbjct: 246 QKARTKWIRERDCNSSYFHKLINYIHRTNAVNGVMIGGSWVEEPNRVKA-VRNFFQIRFS 70
Query: 61 NISGDGVLLEGTTFRSVSEEDN 82
D + G FRS+ ++ N
Sbjct: 69 ESDYDRP*INGANFRSIGQQQN 4
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 73.6 bits (179), Expect = 3e-13
Identities = 29/65 (44%), Positives = 47/65 (71%)
Frame = +2
Query: 444 GGKVCLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCR 503
GG+V LI+SVLS++P+ LSFFK+P+ + +++QFLWGG + +I WVKW+ +C
Sbjct: 5 GGRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICN 184
Query: 504 LRMEG 508
+++G
Sbjct: 185 PKIDG 199
>TC234193
Length = 697
Score = 69.7 bits (169), Expect = 5e-12
Identities = 41/115 (35%), Positives = 69/115 (59%), Gaps = 5/115 (4%)
Frame = -3
Query: 67 VLLEGTTFRSVSEEDNLALTRTFNL--EEIRTAV---WDCEGDKSPGPDGYNFHFIKSFW 121
VLL+G F++++ EDN LT L + R ++ W + D + GYNF+FIK
Sbjct: 326 VLLDGVNFKTINGEDNDRLTHVLVLGTQNKRGSLGV*WKQKFDSN----GYNFNFIKKC* 159
Query: 122 HVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYK 176
V+K+D+VR +++FH++G + N+SF+T + P GL D+ P++L+G + K
Sbjct: 158 EVMKEDVVRAIQEFHSHGCLSRGTNASFLT-----HPP*GLGDFRPVTLVGSLCK 9
>BI698930
Length = 384
Score = 63.2 bits (152), Expect(2) = 4e-10
Identities = 34/71 (47%), Positives = 43/71 (59%)
Frame = +3
Query: 431 KRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGT 490
K L+LWK + LSF G+ CLI S+LSS+PLF SFFK+P V I + F+ G +
Sbjct: 75 K*LALWKHEVLSFAGRTCLINSMLSSLPLFSFSFFKVPLNVGMRFISI*RNFV-GEWRES 251
Query: 491 RKIAWVKWSKV 501
I WV W KV
Sbjct: 252 *MIPWVNWDKV 284
Score = 20.0 bits (40), Expect(2) = 4e-10
Identities = 8/12 (66%), Positives = 9/12 (74%)
Frame = +1
Query: 406 VYLGIHVGANPR 417
VYL I + ANPR
Sbjct: 1 VYLRIPISANPR 36
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 60.8 bits (146), Expect = 2e-09
Identities = 36/112 (32%), Positives = 56/112 (49%)
Frame = -3
Query: 267 CIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFK 326
C+ + ++S+L NG D F + DP+AP+LF++ E LS ++ L + +
Sbjct: 668 CMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKKL*RSIQ 489
Query: 327 VGREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSK 378
V R G+ +S L F DD + E + V + L F SG KVN K+K
Sbjct: 488 VNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTK 333
Score = 37.4 bits (85), Expect = 0.028
Identities = 28/99 (28%), Positives = 47/99 (47%)
Frame = -2
Query: 369 GLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSK 428
G F SK V ED+ ++ L ++T+ YLG+ + T+D ++ K
Sbjct: 345 GQNQKFSFSKNVNWWVEEDI----SSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDK 178
Query: 429 LRKRLSLWKRKTLSFGGKVCLIRSVLSSIPLFYLSFFKL 467
+ KR S WK LS G++ L + V+ ++P + F L
Sbjct: 177 VNKRFSRWKSNLLSMVGRLTLTK*VV*ALPSHIMQFDNL 61
>TC206673
Length = 742
Score = 58.5 bits (140), Expect = 1e-08
Identities = 21/44 (47%), Positives = 34/44 (76%)
Frame = +3
Query: 1 QKSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVVDGGWVEDP 44
QKSRD+W+K+GD+N+ +FH IN+RR N ++G+++ G W+ P
Sbjct: 162 QKSRDKWLKEGDNNSAYFHTVINFRRHYNGLQGILIQGEWIALP 293
>BU762692
Length = 423
Score = 57.4 bits (137), Expect = 3e-08
Identities = 28/59 (47%), Positives = 37/59 (62%)
Frame = -3
Query: 360 MLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGANPRK 418
+LR FEL+SGLK+NF KS + + A+ L+C IPFVY GI +GANPR+
Sbjct: 181 ILRTFELVSGLKINFAKSCFGAFGMTDRWKTEAASCLNCSLLAIPFVYQGIPIGANPRR 5
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 52.0 bits (123), Expect = 1e-06
Identities = 21/46 (45%), Positives = 30/46 (64%)
Frame = +2
Query: 463 SFFKLPKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCRLRMEG 508
SFF LP + + ++++FLWGGE +RKIAWV W VC + +G
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKG 142
>TC234877
Length = 534
Score = 44.7 bits (104), Expect = 2e-04
Identities = 34/135 (25%), Positives = 53/135 (39%), Gaps = 6/135 (4%)
Frame = +1
Query: 394 NLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGGKVCLIRSV 453
N+L +PF+YL + + + P+ +++ L WK LSF G V LI S+
Sbjct: 10 NILGFSVGSLPFIYLSVPLFQEKPRPVHLRPIADRIKAELQSWKGNLLSFMGCVQLIISI 189
Query: 454 LSSIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVC------RLRME 507
+ + L + PK + + F+W K + W K C R +
Sbjct: 190 IDGMLL*SFHLYSWPKALLIEMQTWTRNFIWTRGYFKEKTGYC-WLKNCVPPGMKRALIS 366
Query: 508 GSWESRT*SCLFLPC 522
S *SC F C
Sbjct: 367 LRSLSHK*SCFFETC 411
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 44.7 bits (104), Expect = 2e-04
Identities = 20/36 (55%), Positives = 25/36 (68%)
Frame = +1
Query: 265 KGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPF 300
K C+ SAS+S+LVNGSPT+EF + L G P PF
Sbjct: 1 KECLSSASISILVNGSPTEEFKPXRGLRQGGPFGPF 108
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.148 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,779,572
Number of Sequences: 63676
Number of extensions: 611449
Number of successful extensions: 4494
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 4455
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4490
length of query: 839
length of database: 12,639,632
effective HSP length: 105
effective length of query: 734
effective length of database: 5,953,652
effective search space: 4369980568
effective search space used: 4369980568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0154.9


