

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0151.11
(1562 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
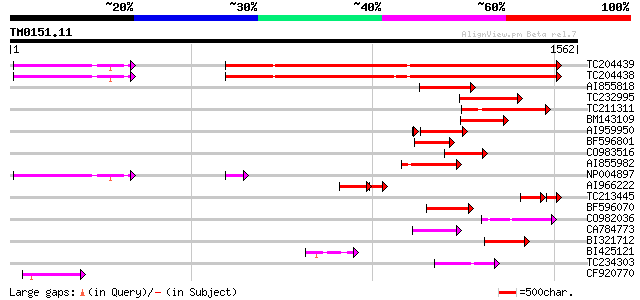
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 879 0.0
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 874 0.0
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 226 6e-59
TC232995 222 9e-58
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 221 3e-57
BM143109 169 9e-42
AI959950 165 2e-40
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 158 2e-38
CO983516 152 1e-36
AI855982 146 6e-35
NP004897 gag-protease polyprotein 136 8e-32
AI966222 113 2e-30
TC213445 88 6e-29
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 123 6e-28
CO982036 123 7e-28
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 114 4e-25
BI321712 110 6e-24
BI425121 110 6e-24
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 108 2e-23
CF920770 107 3e-23
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 879 bits (2270), Expect = 0.0
Identities = 451/931 (48%), Positives = 612/931 (65%), Gaps = 5/931 (0%)
Frame = +1
Query: 595 MLQISLIAPLKHQSWYLDSGCSRHMTGESRMFQELKLKPGGEVGFGGNEKGKIVGTGTIC 654
++ SL A K + WYLDSGCSRHMTG ++ V FG KGKI+G G +
Sbjct: 1645 VVHTSLRASAK-EDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLV 1821
Query: 655 VDSSPCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNI 714
D P ++ VLLV GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N
Sbjct: 1822 HDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNC 2001
Query: 715 YKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDAL 774
Y E + CL S +E +WH+R GH +R + ++ VRG+PNLK +
Sbjct: 2002 YLWTPQETSYSST-CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRI 2178
Query: 775 CEACQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTW 834
C CQ GK K+ + +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TW
Sbjct: 2179 CGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTW 2358
Query: 835 VKFLTRKDESHVVFSTFIAQVQNEKACRIVRVRSDHGGEFES-----LFDSYGIAHDFSC 889
V F+ K E+ VF ++Q EK C I R+RSDHG EFE+ S GI H+FS
Sbjct: 2359 VNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSA 2538
Query: 890 PRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFLAEAVNTACYIQNRISVRPILNKTPY 949
TPQQNG+VERKNRTLQE AR ML + + AEA+NTACYI NR+++R T Y
Sbjct: 2539 AITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLY 2718
Query: 950 ELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKT 1009
E+WK KP++ +FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T
Sbjct: 2719 EIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRT 2898
Query: 1010 IEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEDEPEEEAGPSDSQTL 1069
+ ESI+V DD + + + E NV+D K+ E AE D +E+ +
Sbjct: 2899 VMESINVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAE-NSDSATDESNINQPDKR 3075
Query: 1070 KKSRITAAHPKELILGNKDEPVRTRSAFRPYEETLLSLKGLVSLIEPKSIDEALQDKDWI 1129
+RI HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI
Sbjct: 3076 SSTRIQKMHPKELIIGDPNRGVTTRSR----EVEIVSNSCFVSKIEPKNVKEALTDEFWI 3243
Query: 1130 LAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQE 1189
AM+EEL QF +N+VW LV +PE +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q E
Sbjct: 3244 NAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIE 3423
Query: 1190 GIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPLGFEDE 1249
G+D+ ETFAPVARLE+IRLL+ + L+QMDVKSAFLNGY++EEVYV QP GF D
Sbjct: 3424 GVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADP 3603
Query: 1250 KKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIY 1309
PDHV++LKK+LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIY
Sbjct: 3604 THPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIY 3783
Query: 1310 VDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELL 1369
VDDI+FG + + + F + MQ+EFEMS++GEL YFLG+QV Q + ++ QS+Y K ++
Sbjct: 3784 VDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIV 3963
Query: 1370 KKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCA 1429
KKF M ++ +TP L K++ V Q LYR MIGSLLYLTASRPDI ++V +CA
Sbjct: 3964 KKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCA 4143
Query: 1430 RFQSDPRETHLTAIKRILRYLKGTTNLGLMYKKTSEYKLSGYCDAHYAGDRTERKSTSGN 1489
R+Q++P+ +HLT +KRIL+Y+ GT++ G+MY S L GYCDA +AG +RKSTSG
Sbjct: 4144 RYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGG 4323
Query: 1490 CQFLGSNLVSWASKRQSTIALSTAEAEYISA 1520
C +LG+NL+SW SK+Q+ ++LSTAEAEYI+A
Sbjct: 4324 CFYLGNNLISWFSKKQNCVSLSTAEAEYIAA 4416
Score = 140 bits (352), Expect = 6e-33
Identities = 97/350 (27%), Positives = 164/350 (46%), Gaps = 15/350 (4%)
Frame = +1
Query: 11 KPPMFDGQRFEYWKDRMESFFLGFDADLWDIIVDGYERP--VDADGKKI----PRSEMTA 64
+PP+ DG +EYWK RM +F D+ W ++ G+E P +D +GK P + T
Sbjct: 34 RPPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTK 213
Query: 65 DQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFESLKMSHEGNKKVKESKALSLI 124
++ +L + KA L + + ++ I AK +E LK++HEG KVK S+ L
Sbjct: 214 EEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLA 393
Query: 125 QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRCLPESWMPLVTSIE 184
K+E+ M+ E I + + L + T + V +++R LP+ + VT+IE
Sbjct: 394 TKFENLKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIE 573
Query: 185 LTRDVENMSLEELISILKCHELKRSEMQDLRKKSIALKSKSEKAKAEKSKALQAEEEESE 244
+D+ NM ++ELI L+ EL S+ + + K++A S E EE+E +
Sbjct: 574 EAQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSNDE-----------GEEDEYD 720
Query: 245 EASEDSDEDELTLISKRLNRIWKHRQSKYK--------GSGKAKGKSESSGQKKSSLKEV 296
+++ + + L+ K+ N++ + K K + S K S K +
Sbjct: 721 LDTDEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGI 900
Query: 297 TCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMVTFDESESE-DVDSD 345
C C+ GH ++CP K K +K L V ++ESE + DSD
Sbjct: 901 QCHGCEGYGHIIAECPTHLK--------KHRKGLSVCQSDTESEQESDSD 1026
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 874 bits (2257), Expect = 0.0
Identities = 451/933 (48%), Positives = 612/933 (65%), Gaps = 7/933 (0%)
Frame = +1
Query: 595 MLQISLIAPLKHQSWYLDSGCSRHMTGESRMFQELKLKPGGEVGFGGNEKGKIVGTGTIC 654
++ SL A K + WYLDSGCSRHMTG ++ V FG KGKI G G +
Sbjct: 1648 VVHTSLRASAK-EDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLV 1824
Query: 655 VDSSPCIDNVLLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNI 714
D P ++ VLLV GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N
Sbjct: 1825 HDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNC 2004
Query: 715 YKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDAL 774
Y E + CL S +E +WH+R GH +R + ++ VRG+PNLK +
Sbjct: 2005 YLWTPQETSYSST-CLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRI 2181
Query: 775 CEACQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTW 834
C CQ GK K+ + +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TW
Sbjct: 2182 CGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTW 2361
Query: 835 VKFLTRKDESHVVFSTFIAQVQNEKACRIVRVRSDHGGEFES-----LFDSYGIAHDFSC 889
V F+ K ++ VF ++Q EK C I R+RSDHG EFE+ S GI H+FS
Sbjct: 2362 VNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSA 2541
Query: 890 PRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFLAEAVNTACYIQNRISVRPILNKTPY 949
TPQQNG+VERKNRTLQE AR ML + + AEA+NTACYI NR+++R T Y
Sbjct: 2542 AITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLY 2721
Query: 950 ELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSERSKGFRFYNTDAKT 1009
E+WK KP + +FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T
Sbjct: 2722 EIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRT 2901
Query: 1010 IEESIHVRFDDKLDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEDEPEEEAGPSDSQTL 1069
+ ESI+V DD + + + E NV+D K+ E AE + +E P+ +Q
Sbjct: 2902 VMESINVVVDDLTPARKKDVEEDVRTSGDNVADTAKSAENAENSDSATDE---PNINQPD 3072
Query: 1070 KKS--RITAAHPKELILGNKDEPVRTRSAFRPYEETLLSLKGLVSLIEPKSIDEALQDKD 1127
K+ RI HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+
Sbjct: 3073 KRPSIRIQKMHPKELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEPKNVKEALTDEF 3240
Query: 1128 WILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQ 1187
WI AM+EEL QF +N+VW LV +PE +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q
Sbjct: 3241 WINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQ 3420
Query: 1188 QEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPLGFE 1247
EG+D+ ETFAPVARLE+IRLL+ + L+QMDVKSAFLNGY++EE YV QP GF
Sbjct: 3421 IEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFV 3600
Query: 1248 DEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQ 1307
D PDHV++LKK+LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I Q
Sbjct: 3601 DPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQ 3780
Query: 1308 IYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKE 1367
IYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+QV Q + ++ QSKY K
Sbjct: 3781 IYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKN 3960
Query: 1368 LLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDILFSVHL 1427
++KKF M ++ +TP L K++ V Q LYR MIGSLLYLTASRPDI ++V +
Sbjct: 3961 IVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGV 4140
Query: 1428 CARFQSDPRETHLTAIKRILRYLKGTTNLGLMYKKTSEYKLSGYCDAHYAGDRTERKSTS 1487
CAR+Q++P+ +HL +KRIL+Y+ GT++ G+MY S+ L GYCDA +AG +RKSTS
Sbjct: 4141 CARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKSTS 4320
Query: 1488 GNCQFLGSNLVSWASKRQSTIALSTAEAEYISA 1520
G C +LG+NL+SW SK+Q+ ++LSTAEAEYI+A
Sbjct: 4321 GGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAA 4419
Score = 142 bits (357), Expect = 1e-33
Identities = 99/351 (28%), Positives = 166/351 (47%), Gaps = 16/351 (4%)
Frame = +1
Query: 11 KPPMFDGQRFEYWKDRMESFFLGFDADLWDIIVDGYERP--VDADGKKI----PRSEMTA 64
+PP+ DG +EYWK RM +F D+ W ++ G+E P +D +GK P + T
Sbjct: 34 RPPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTK 213
Query: 65 DQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFESLKMSHEGNKKVKESKALSLI 124
++ +L + KA L + + ++ I AK +E LK +HEG KVK S+ L
Sbjct: 214 EEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLA 393
Query: 125 QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRCLPESWMPLVTSIE 184
K+E+ M+ E I + + L + T + V +++R LP+ + VT+IE
Sbjct: 394 TKFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIE 573
Query: 185 LTRDVENMSLEELISILKCHELKRSEMQDLRKKSIALKSKSEKAKAEKSKALQAEEEESE 244
+D+ NM ++ELI L+ EL S+ + + K++A S E EE+E +
Sbjct: 574 EAQDICNMRVDELIGSLQTFELGLSDRTEKKSKNLAFVSNDE-----------GEEDEYD 720
Query: 245 EASEDSDEDELTLISKRLNRIWKHRQSKYK--------GSGKAKGKSESSGQKKSSLKEV 296
+++ + + L+ K+ N++ + K K + S +K S K +
Sbjct: 721 LDTDEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKKSDEKPSHSKGI 900
Query: 297 TCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMV-TFDESESE-DVDSD 345
C C+ GH K++CP K K +K L V D++ESE + DSD
Sbjct: 901 QCHGCEGYGHIKAECPTHLK--------KQRKGLSVCRSDDTESEQESDSD 1029
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 226 bits (576), Expect = 6e-59
Identities = 108/153 (70%), Positives = 131/153 (85%)
Frame = -3
Query: 1130 LAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQE 1189
+AM+EELNQF +N+VW LV+KPE+ VIGTKWVFRNKL+E G ++RNKARLVA+GY+Q+E
Sbjct: 461 IAMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEE 282
Query: 1190 GIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPLGFEDE 1249
GIDY ET+APVARLE IR+L+++ N L+QMDVKSAFLNG I EEVYV QP GFE
Sbjct: 281 GIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIP 102
Query: 1250 KKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLE 1282
KP HV+KL+K+LYGLKQAPRAWYER+S+FLLE
Sbjct: 101 DKPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>TC232995
Length = 1009
Score = 222 bits (566), Expect = 9e-58
Identities = 111/173 (64%), Positives = 132/173 (76%)
Frame = +2
Query: 1240 VHQPLGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTY 1299
V QP GFE KP+HV+KL+K+LYGLKQAPRAWYERLS+FLLE EF RGKVDTTLF K
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 1300 KDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYI 1359
+DIL+VQIYVDDIIFGS N SLCKEFS MQ+EFEMSMMGELKYFLG+Q+ QT G +I
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 1360 HQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLL 1412
+QSKY KEL+K+F M + TPM C L+K++ + K YR IG ++
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVV 520
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 221 bits (562), Expect = 3e-57
Identities = 121/247 (48%), Positives = 160/247 (63%), Gaps = 2/247 (0%)
Frame = +3
Query: 1245 GFEDEKKPDHVFKLKKSL--YGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDD 1302
GFED+++P HVF + L G+K ++ S ++ K+
Sbjct: 330 GFEDKERPCHVFMV*NKL*ELGMKG*VHF*FQMDSPEE*RTPHYSERLK--------KET 485
Query: 1303 ILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQS 1362
LI+ IYVDDIIFG+ ++ +CKEF E+M+ FE SM GELK+ LG+Q+ Q G +IHQ
Sbjct: 486 FLIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIFIHQE 665
Query: 1363 KYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDIL 1422
KYTK LK+F M E+ TPMH + I++K++K K Y GMI SL YLT+SRPDI+
Sbjct: 666 KYTKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTSSRPDIV 845
Query: 1423 FSVHLCARFQSDPRETHLTAIKRILRYLKGTTNLGLMYKKTSEYKLSGYCDAHYAGDRTE 1482
F V LCARFQS P+ +H+TA+KRILRYL GTTN L +KK SE+ L GYCD ++AGD+ E
Sbjct: 846 FVVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYFAGDKVE 1025
Query: 1483 RKSTSGN 1489
RKSTS N
Sbjct: 1026 RKSTSRN 1046
>BM143109
Length = 415
Score = 169 bits (428), Expect = 9e-42
Identities = 85/133 (63%), Positives = 102/133 (75%)
Frame = +1
Query: 1242 QPLGFEDEKKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD 1301
QP ++ +KP+HVFKLKK LYGLKQA RAWYE LS FLL+ F +GKVDT LF +
Sbjct: 4 QPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLN 183
Query: 1302 DILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1361
DIL+VQIYVDDIIFGS N SLCK+FS+ MQ EFEMSMM EL +FLG+Q+ QT G +I Q
Sbjct: 184 DILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFISQ 363
Query: 1362 SKYTKELLKKFNM 1374
SKY K+L+ +F M
Sbjct: 364 SKYCKDLIHRFGM 402
>AI959950
Length = 466
Score = 165 bits (417), Expect(2) = 2e-40
Identities = 85/130 (65%), Positives = 101/130 (77%)
Frame = -1
Query: 1131 AMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEG 1190
AM+EEL+QF KN+V LVK P+ V+G KW+F NKL+E G VVR KARLVA+GYSQQEG
Sbjct: 391 AMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEG 212
Query: 1191 IDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPLGFEDEK 1250
IDY +TFA VARLE I +L+SF+ N+ L+QMDVKSAFLNG I +EVYV QP GFE+E
Sbjct: 211 IDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFENET 32
Query: 1251 KPDHVFKLKK 1260
HVFKL K
Sbjct: 31 LHQHVFKLNK 2
Score = 21.2 bits (43), Expect(2) = 2e-40
Identities = 8/16 (50%), Positives = 13/16 (81%)
Frame = -2
Query: 1110 LVSLIEPKSIDEALQD 1125
L+ ++PK IDEA++D
Sbjct: 453 LIFEMKPKHIDEAIKD 406
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 158 bits (399), Expect = 2e-38
Identities = 73/111 (65%), Positives = 95/111 (84%)
Frame = +3
Query: 1115 EPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVV 1174
EPK+I EA+ D +WI+ M+EELNQF +N+VW LV+KPE+ VIGTKWVFRNKL+E G ++
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 1175 RNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDV 1225
RNKARLVA+GY+Q+EGIDY ET+APVARLEAIR+L++++ N L+QMDV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>CO983516
Length = 724
Score = 152 bits (384), Expect = 1e-36
Identities = 73/120 (60%), Positives = 92/120 (75%)
Frame = +2
Query: 1197 FAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPLGFEDEKKPDHVF 1256
F PVARLE+IRLL+ + L+QMDVKSAFLNGY++EEVYV QP GF D PDHV+
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 1257 KLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFG 1316
+LKK+LYGLKQAPRAWYERL+ L + + +G +D TLF K ++++I QIYVDDI+FG
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFG 724
>AI855982
Length = 484
Score = 146 bits (369), Expect = 6e-35
Identities = 77/165 (46%), Positives = 111/165 (66%)
Frame = +2
Query: 1079 PKELILGNKDEPVRTRSAFRPYEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEELNQ 1138
P + I+G+ + V TR + + L + VS+IEPK+I EA+ D +WI+AM+EELNQ
Sbjct: 2 PLDNIIGDISKGVTTRHSLKD----LCNNMAFVSMIEPKNIKEAIVDDNWIIAMQEELNQ 169
Query: 1139 FSKNDVWSLVKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFA 1198
F +N+VW LV+KP++ VI TKWVFRNKL+E ++ +KARLVA+GY+Q +G+DY T+A
Sbjct: 170 FERNNVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIIIHKARLVAEGYNQVDGLDYEHTYA 349
Query: 1199 PVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQP 1243
+ARL I + +S+ N L+ SA L+G + EVYV QP
Sbjct: 350 SIARL*VIIMPLSYVYIMNSTLYHYACVSALLHGLLLHEVYVDQP 484
>NP004897 gag-protease polyprotein
Length = 1923
Score = 136 bits (342), Expect = 8e-32
Identities = 97/351 (27%), Positives = 164/351 (46%), Gaps = 16/351 (4%)
Frame = +1
Query: 11 KPPMFDGQRFEYWKDRMESFFLGFDADLWDIIVDGYERP--VDADGKKI----PRSEMTA 64
+PP+ DG +EYWK RM +F D+ W ++ +E P +D +GK P + T
Sbjct: 34 RPPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKDWEHPKMLDTEGKPTDGLKPEEDWTK 213
Query: 65 DQKKLYSQHHKARAILLSAISYEEYQKITDREFAKGIFESLKMSHEGNKKVKESKALSLI 124
++ +L + KA L + + ++ I AK +E LK +HEG KVK S+ L
Sbjct: 214 EEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLA 393
Query: 125 QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRCLPESWMPLVTSIE 184
K+E+ M+ E I + + L + T + V +++R LP+ + VT+IE
Sbjct: 394 TKFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIE 573
Query: 185 LTRDVENMSLEELISILKCHELKRSEMQDLRKKSIALKSKSEKAKAEKSKALQAEEEESE 244
+D+ N+ ++ELI L+ EL S+ + + K++A S E EE+E +
Sbjct: 574 EAQDICNLRVDELIGSLQTFELGLSDRTEKKSKNLAFVSNDE-----------GEEDEYD 720
Query: 245 EASEDSDEDELTLISKRLNRIWKHRQSKYK--------GSGKAKGKSESSGQKKSSLKEV 296
+++ + + L+ K+ N++ + K K + S +K S K
Sbjct: 721 LDTDEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKRSDEKPSHSKGF 900
Query: 297 TCFECKESGHYKSDCPKLKKDKKPKKHFKTKKSLMV-TFDESESE-DVDSD 345
C C+ GH K++CP K K +K L V D++ESE + DSD
Sbjct: 901 QCHGCEGYGHIKAECPTHLK--------KQRKGLSVCRSDDTESEQESDSD 1029
Score = 50.1 bits (118), Expect = 8e-06
Identities = 27/62 (43%), Positives = 33/62 (52%)
Frame = +1
Query: 595 MLQISLIAPLKHQSWYLDSGCSRHMTGESRMFQELKLKPGGEVGFGGNEKGKIVGTGTIC 654
++ SL A K + WYLDSGCSRHMTG ++ V FG KGKI G G +
Sbjct: 1648 VVHTSLRASAK-EDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLV 1824
Query: 655 VD 656
D
Sbjct: 1825 HD 1830
>AI966222
Length = 430
Score = 113 bits (283), Expect(2) = 2e-30
Identities = 50/88 (56%), Positives = 64/88 (71%)
Frame = +1
Query: 908 EMARTMLQETGMAKHFLAEAVNTACYIQNRISVRPILNKTPYELWKNIKPNISYFHPFGC 967
EMART L + KHF AE +N CY+QN+I +RPIL +TPYELWK KPNISYF+PF C
Sbjct: 1 EMARTTLNDNLTPKHF*AEVMNIVCYLQNKIYIRPILKRTPYELWKGRKPNISYFYPFRC 180
Query: 968 VCYVLNTKDRLHKFDAKSSKCLLLGYSE 995
C+++NTKD L K D+KS + + YS+
Sbjct: 181 KCFIINTKDNLGKIDSKSDCGIFIAYSK 264
Score = 39.3 bits (90), Expect(2) = 2e-30
Identities = 22/51 (43%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Frame = +2
Query: 990 LLGYSERSKGFRFYNTDAKTIEESIHVRF-DDKLDSDQSKLVEKFADLSIN 1039
LL + SK FR YN+ IEE+IH+RF +K + + +L E FADL ++
Sbjct: 248 LLHTLKLSKAFRVYNSGTLVIEEAIHIRFGKNKPNKELLELDESFADLRLD 400
>TC213445
Length = 705
Score = 87.8 bits (216), Expect(2) = 6e-29
Identities = 41/68 (60%), Positives = 51/68 (74%)
Frame = +2
Query: 1407 MIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAIKRILRYLKGTTNLGLMYKKTSEY 1466
MI S LYL+ SRP I+FSV +C R+Q++P+E+HL+ IKRI+RYL G NLGL Y K S Y
Sbjct: 197 MIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPKNSSY 376
Query: 1467 KLSGYCDA 1474
L GY DA
Sbjct: 377 NLVGYSDA 400
Score = 60.1 bits (144), Expect(2) = 6e-29
Identities = 27/41 (65%), Positives = 35/41 (84%)
Frame = +1
Query: 1480 RTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISA 1520
+T+R+STS C F+GS LVSW SK+Q+++ LSTAEAEYISA
Sbjct: 400 KTDRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISA 522
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 123 bits (309), Expect = 6e-28
Identities = 60/130 (46%), Positives = 89/130 (68%)
Frame = -2
Query: 1148 VKKPESVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIR 1207
V P +G +WV+ K+ G+V R KARLVA+GY+Q GIDY +TF+PVA+L +R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 1208 LLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPLGFEDEKKPDHVFKLKKSLYGLKQ 1267
L ++ + + LHQ+D+K+AFL+G + E++Y+ QP GF + + V KL +SLYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 1268 APRAWYERLS 1277
+PRAW+ + S
Sbjct: 46 SPRAWFGKFS 17
>CO982036
Length = 674
Score = 123 bits (308), Expect = 7e-28
Identities = 74/212 (34%), Positives = 118/212 (54%), Gaps = 5/212 (2%)
Frame = -2
Query: 1299 YKDDILIVQ--IYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEG 1356
YK IL V +YVD II GS+ +L + + + + F + ++G+L YF+ I+V P+
Sbjct: 673 YKTHILTVYLLVYVDIIITGSSC-TLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDL 497
Query: 1357 TYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTA 1416
+ ++ + +K ++ +PM TC L K D YR ++G+L Y T
Sbjct: 496 LFSLRTSIFEIFCRKPR*QAQPIS-SPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTV 320
Query: 1417 SRPDILFSVHLCARFQSDPRETHLTAIKRILRYLKGTTNLGLMYK---KTSEYKLSGYCD 1473
RP+I F+V+ +F S+P ++H T +KRILRYLKG+ + GL K + + G+CD
Sbjct: 319 IRPEISFAVNKVCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCD 140
Query: 1474 AHYAGDRTERKSTSGNCQFLGSNLVSWASKRQ 1505
A +A +++STSG FLG NL+SW +Q
Sbjct: 139 ADWASAVDDKRSTSGAAVFLGPNLISWWXXKQ 44
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 114 bits (284), Expect = 4e-25
Identities = 56/134 (41%), Positives = 81/134 (59%)
Frame = +3
Query: 1110 LVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPESVHVIGTKWVFRNKLNE 1169
L SL P +I EAL W AM +E+ N W LV P +G +WV+ K+
Sbjct: 3 LSSLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGP 182
Query: 1170 KGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAF 1229
G V R KARLVA+GY+Q GI+Y +TF+PV L +RL ++ + + LHQ+D+K+AF
Sbjct: 183 NGKVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAF 362
Query: 1230 LNGYISEEVYVHQP 1243
L+G + E++Y+ QP
Sbjct: 363 LHGDLEEDIYMEQP 404
>BI321712
Length = 399
Score = 110 bits (274), Expect = 6e-24
Identities = 57/124 (45%), Positives = 79/124 (62%)
Frame = -3
Query: 1308 IYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKE 1367
+YVDD+IF N S+ +EF + M EFEM+ MG + Y+LGI+V Q +G +I Q Y KE
Sbjct: 379 LYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQEDKGIFITQEGYAKE 200
Query: 1368 LLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDILFSVHL 1427
+LKKF M ++ TPM L K +K V LY+ +IGSL YLT +RPDIL+ V +
Sbjct: 199 VLKKFKMDDANPVGTPMECGSKLSKHEKGENVDPTLYKSLIGSLRYLTCTRPDILYVVGV 20
Query: 1428 CARF 1431
+R+
Sbjct: 19 VSRY 8
>BI425121
Length = 412
Score = 110 bits (274), Expect = 6e-24
Identities = 67/157 (42%), Positives = 86/157 (54%), Gaps = 10/157 (6%)
Frame = +3
Query: 815 SIGGKRYGMVIVDDYSRWTWVKFLTRKDES----------HVVFSTFIAQVQNEKACRIV 864
S+G K+Y + VDDYSR+TWV FL K ES VF + +V E + RI+
Sbjct: 6 SLGCKKYEFLTVDDYSRYTWVYFLAHKHESLRYFIRGFKMKKVFVFLLLEVTMELSLRIL 185
Query: 865 RVRSDHGGEFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFL 924
+ GI H+ S PRTPQ+N VVERKNRTLQE ART+L
Sbjct: 186 --------S*NHFCERNGIFHNLS*PRTPQENRVVERKNRTLQEKARTIL---------- 311
Query: 925 AEAVNTACYIQNRISVRPILNKTPYELWKNIKPNISY 961
T C++QN+I +RP++ KTPYELWK + ISY
Sbjct: 312 ----FTTCFVQNKILIRPMIKKTPYELWKGRRHIISY 410
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 108 bits (269), Expect = 2e-23
Identities = 69/182 (37%), Positives = 105/182 (56%), Gaps = 4/182 (2%)
Frame = +1
Query: 1171 GDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFL 1230
G + + KARLVA+ Y+Q G DYT TF+PVA++ + LL S +V + L +D K+AFL
Sbjct: 28 GTIDQFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFL 207
Query: 1231 NGYISEEVYVHQPLGFEDE-KKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFV--- 1286
+GY+ EEVY+ QPLGF + + + V +L +S YGLKQ+PRAW FL +
Sbjct: 208 HGYLEEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAW-----PFLYCGAAIWYD 372
Query: 1287 RGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFL 1346
+ D ++F + + +YVDDI ++Q + +F+ +G+L+YFL
Sbjct: 373 SHEADHSVFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFL 552
Query: 1347 GI 1348
GI
Sbjct: 553 GI 558
>CF920770
Length = 581
Score = 107 bits (268), Expect = 3e-23
Identities = 61/186 (32%), Positives = 102/186 (54%), Gaps = 13/186 (6%)
Frame = -2
Query: 35 DADLWDIIVDGYERP-----VDADGKKI--------PRSEMTADQKKLYSQHHKARAILL 81
D ++W+ I G P V DG PR + + +K + KA+ I+
Sbjct: 574 DLNIWEAIEIGPYIPTTVERVSIDGSSSSESITIEKPRDRWSEEDRKRVQYNLKAKNIIT 395
Query: 82 SAISYEEYQKITDREFAKGIFESLKMSHEGNKKVKESKALSLIQKYESFIMEPNESIEEM 141
SA+ +EY ++++ + AK ++++L+++HEG VK S+ +L +YE F M NE+I+ M
Sbjct: 394 SALGMDEYFRVSNCKSAKEMWDTLRLTHEGTTDVKRSRINALTHEYELFRMNTNENIQSM 215
Query: 142 FSRFQLLVAGIRPLNKSYTTKDHVIRVIRCLPESWMPLVTSIELTRDVENMSLEELISIL 201
RF +V + L K + +D + +V+RCL W P VT+I +RD+ NMSL L L
Sbjct: 214 QKRFTHIVNHLAALGKEFQNEDLINKVLRCLSREWQPKVTAISESRDLSNMSLATLFGKL 35
Query: 202 KCHELK 207
+ HE++
Sbjct: 34 QEHEME 17
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,981,729
Number of Sequences: 63676
Number of extensions: 903933
Number of successful extensions: 5100
Number of sequences better than 10.0: 174
Number of HSP's better than 10.0 without gapping: 4872
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5037
length of query: 1562
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1452
effective length of database: 5,635,272
effective search space: 8182414944
effective search space used: 8182414944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0151.11


