

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0148.5
(586 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
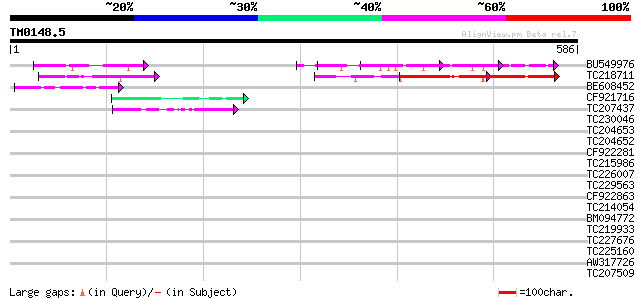
Score E
Sequences producing significant alignments: (bits) Value
BU549976 123 3e-28
TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), ... 120 1e-27
BE608452 52 1e-06
CF921716 45 1e-04
TC207437 similar to UP|O81922 (O81922) Proline-rich protein, par... 43 3e-04
TC230046 similar to UP|Q9LU79 (Q9LU79) Gb|AAF21150.1, partial (28%) 42 0.001
TC204653 similar to UP|Q9AT20 (Q9AT20) Histone H1 (Fragment), pa... 40 0.004
TC204652 similar to UP|H1_PEA (P08283) Histone H1, partial (81%) 39 0.005
CF922281 39 0.005
TC215986 similar to GB|AAC06384.1|1732361|MDU80269 translation i... 38 0.011
TC226007 similar to UP|Q8LC13 (Q8LC13) Remorin, partial (65%) 38 0.011
TC229563 38 0.011
CF922863 38 0.014
TC214054 similar to UP|Q75QC2 (Q75QC2) Glutamate-rich protein, p... 38 0.014
BM094772 37 0.019
TC219933 weakly similar to UP|Q9EQF1 (Q9EQF1) GABA-A epsilon sub... 37 0.019
TC227676 37 0.019
TC225160 UP|Q948X9 (Q948X9) Beta-conglycinin alpha-subunit, comp... 37 0.025
AW317726 weakly similar to PIR|T17104|T171 translation initiatio... 37 0.032
TC207509 37 0.032
>BU549976
Length = 735
Score = 123 bits (308), Expect = 3e-28
Identities = 90/227 (39%), Positives = 121/227 (52%), Gaps = 22/227 (9%)
Frame = -3
Query: 363 NKCNAKLPETLMLASNCHI-----DQAIPSDSN------PAVPTNPEHKTPKKWKKRAKK 411
NK +LPE + S C I D AIP S PA+ +PE + KK KKR
Sbjct: 724 NKNCTELPEISIQTSPCLIEASXXDPAIPMGSCLTXXICPAISADPEQEMSKKKKKR--- 554
Query: 412 KNVLLSEGGKSNEHN----GQPTETPVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSE 467
K+VL SE + NEHN P + PVQKS+ I+APSIDP +P P+D TP
Sbjct: 553 KSVLKSEAAEPNEHNKLPEAAPPDAPVQKSIYHIDAPSIDPIIPMGPSPPVDTETPIDQG 374
Query: 468 GKMSNAQRNN----VLLSEGVKSNE---QPTETPVQKSVNPIKAPSIDPTLPTDQPVPID 520
+M+ +R V+++EG ++NE +P+ETPV++ P A SIDP +P P PID
Sbjct: 373 QEMTKKKRKKKKRIVIITEGTETNEHGVKPSETPVERFTYPTVATSIDPKIPVLAP-PID 197
Query: 521 LATPKHSEGKMSKAQRRKKNKRALKSAWSESVHHSSDQNAKTPVQRT 567
ATP + K K +K+ AL S +E H D+ A TPVQR+
Sbjct: 196 TATPINQSQKTGK---KKRKMSALISKGAEPNEH-DDKPAGTPVQRS 68
Score = 103 bits (257), Expect = 2e-22
Identities = 71/201 (35%), Positives = 107/201 (52%), Gaps = 9/201 (4%)
Frame = -3
Query: 319 DTEQKINKRSRMSKMKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASN 378
D EQ+++K+ + K K + P++HN P E +A + +++
Sbjct: 592 DPEQEMSKKKKKRKSVLKSEAAEPNEHNKLP-----------EAAPPDAPVQKSIYHIDA 446
Query: 379 CHIDQAIPSDSNPAVPTNP-----EHKTPKKWKKRAKKKNVLLSEGGKSNEHNGQPTETP 433
ID IP +P V T + T KK KK KK+ V+++EG ++NEH +P+ETP
Sbjct: 445 PSIDPIIPMGPSPPVDTETPIDQGQEMTKKKRKK--KKRIVIITEGTETNEHGVKPSETP 272
Query: 434 VQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSEGKMSNAQRN-NVLLSEGVKSNE---Q 489
V++ P A SIDP +P P PID ATP + K +R + L+S+G + NE +
Sbjct: 271 VERFTYPTVATSIDPKIPVLAP-PIDTATPINQSQKTGKKKRKMSALISKGAEPNEHDDK 95
Query: 490 PTETPVQKSVNPIKAPSIDPT 510
P TPVQ+S++ I PSIDPT
Sbjct: 94 PAGTPVQRSIHSIAEPSIDPT 32
Score = 67.4 bits (163), Expect = 2e-11
Identities = 52/162 (32%), Positives = 76/162 (46%), Gaps = 9/162 (5%)
Frame = -3
Query: 297 PVIPVCPNSASLIGQSQTIPVVDTEQKINKRSRMSKMKRKRMNSM--------PDKHNTE 348
P+IP+ P+ P VDTE I++ M+K KRK+ + ++H +
Sbjct: 433 PIIPMGPS-----------PPVDTETPIDQGQEMTKKKRKKKKRIVIITEGTETNEHGVK 287
Query: 349 PPETK-KRVLDREEPNKCNAKLPETLMLASNCHIDQAIPSDSNPAVPTNPEHKTPKKWKK 407
P ET +R + K+P A P D+ A P N KT KK
Sbjct: 286 PSETPVERFTYPTVATSIDPKIPVL-----------APPIDT--ATPINQSQKTGKK--- 155
Query: 408 RAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPSIDPT 449
+K + L+S+G + NEH+ +P TPVQ+S+ I PSIDPT
Sbjct: 154 -KRKMSALISKGAEPNEHDDKPAGTPVQRSIHSIAEPSIDPT 32
Score = 44.3 bits (103), Expect = 2e-04
Identities = 39/144 (27%), Positives = 59/144 (40%), Gaps = 25/144 (17%)
Frame = -3
Query: 25 PSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPE--------------------- 63
P+ D EQ SKKK+++K +K E E +EH++ LPE
Sbjct: 607 PAISADPEQEMSKKKKKRKSVLKSEAAEPNEHNK--LPEAAPPDAPVQKSIYHIDAPSID 434
Query: 64 PVV-VATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEP- 121
P++ + + P+ ETP +QG Q K RKKKK+ ++ E
Sbjct: 433 PIIPMGPSPPVDTETPIDQG--------------QEMTKKKRKKKKRIVIITEGTETNEH 296
Query: 122 --EPKETSNTDHSEPTQLCSVKKK 143
+P ET + PT S+ K
Sbjct: 295 GVKPSETPVERFTYPTVATSIDPK 224
>TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), partial (6%)
Length = 1204
Score = 120 bits (302), Expect = 1e-27
Identities = 69/168 (41%), Positives = 101/168 (60%), Gaps = 3/168 (1%)
Frame = -3
Query: 404 KWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPSIDPTLPTDQPVPIDLATP 463
K K++ KK+ V++ EG ++NEH +P ET VQK P SIDPT+P P PID
Sbjct: 1130 KKKRKKKKRIVIIIEGTETNEHGVKPPETLVQKFAYPTVTTSIDPTIPVPAP-PID---- 966
Query: 464 KHSEGKMSNAQRNNVLLSEGVKSNE---QPTETPVQKSVNPIKAPSIDPTLPTDQPVPID 520
+ ++ + L+S+G + +E +P TPVQ+S++PI PSIDPT+P Q P+D
Sbjct: 965 -QGQKMRKKMRKRSALISKGAEPDEHDDKPPGTPVQRSIHPIAEPSIDPTIPASQTPPVD 789
Query: 521 LATPKHSEGKMSKAQRRKKNKRALKSAWSESVHHSSDQNAKTPVQRTE 568
ATP E K +RKK + + K+ ES H++D+ A+TPVQ+TE
Sbjct: 788 PATPVDPEPMTKK--KRKKKRNSAKNEGLESEKHNADRIAETPVQKTE 651
Score = 99.8 bits (247), Expect = 3e-21
Identities = 68/196 (34%), Positives = 104/196 (52%), Gaps = 13/196 (6%)
Frame = -3
Query: 316 PVVDTEQKINKRSRMSKMKRKRMNSMPDKHNTEPPETKKRV---LDREEPNKCNAKLPET 372
P VDTE I++ +M+K KRK+ KKR+ ++ E N+ K PET
Sbjct: 1178 PPVDTETPIDQGQQMTKKKRKK---------------KKRIVIIIEGTETNEHGVKPPET 1044
Query: 373 LMLASNCHIDQAIPSDSNPAVPTNPEHKTP----KKWKKRAKKKNVLLSEGGKSNEHNGQ 428
L+ A P+ + PT P P +K +K+ +K++ L+S+G + +EH+ +
Sbjct: 1043 LVQKF------AYPTVTTSIDPTIPVPAPPIDQGQKMRKKMRKRSALISKGAEPDEHDDK 882
Query: 429 PTETPVQKSVSPINAPSIDPTLPTDQPVPIDLATPKHSE--GKMSNAQRNNVLLSEGVKS 486
P TPVQ+S+ PI PSIDPT+P Q P+D ATP E K ++ N +EG++S
Sbjct: 881 PPGTPVQRSIHPIAEPSIDPTIPASQTPPVDPATPVDPEPMTKKKRKKKRNSAKNEGLES 702
Query: 487 N----EQPTETPVQKS 498
++ ETPVQK+
Sbjct: 701 EKHNADRIAETPVQKT 654
Score = 45.1 bits (105), Expect = 9e-05
Identities = 45/157 (28%), Positives = 67/157 (42%), Gaps = 31/157 (19%)
Frame = -3
Query: 30 DQEQSCSKKKRRKKKKIK--EEGIESHEHDQVLLPEPVVVATAIPITLETPKEQGAEPIE 87
DQ Q +KKKR+KKK+I EG E++EH V PE +V A P T+ T I+
Sbjct: 1151 DQGQQMTKKKRKKKKRIVIIIEGTETNEHG-VKPPETLVQKFAYP-TVTT-------SID 999
Query: 88 ATVVHPVPDQRPAKTHRKKKKKETVL---------------------------EPQPEPE 120
T+ P P + RKK +K + L EP +P
Sbjct: 998 PTIPVPAPPIDQGQKMRKKMRKRSALISKGAEPDEHDDKPPGTPVQRSIHPIAEPSIDPT 819
Query: 121 PEPKETSNTDHSEPT--QLCSVKKKKKRKGAESNEAV 155
+T D + P + + KK+KK++ + NE +
Sbjct: 818 IPASQTPPVDPATPVDPEPMTKKKRKKKRNSAKNEGL 708
>BE608452
Length = 426
Score = 51.6 bits (122), Expect = 1e-06
Identities = 38/112 (33%), Positives = 59/112 (51%)
Frame = +3
Query: 6 QSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPV 65
QSE +KKRR E+ P+ L S D E+ K+KR++KKKI+ ES+E QV +
Sbjct: 60 QSEFSDAKKRREEDNPQTLVSLVHDSEK--KKRKRKRKKKIE----ESNEQPQVEVCPKS 221
Query: 66 VVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQP 117
+V + IPI + KE E + +P P ++ K ++ + E +L P
Sbjct: 222 LVDSVIPIVAQ--KENAETTTENS--NPTPKKKKKKKNKNVQDAEQLLTCDP 365
Score = 38.1 bits (87), Expect = 0.011
Identities = 33/112 (29%), Positives = 49/112 (43%), Gaps = 15/112 (13%)
Frame = +3
Query: 79 KEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKE------------- 125
K++ E T+V V D K RK+KKK QP+ E PK
Sbjct: 81 KKRREEDNPQTLVSLVHDSEKKKRKRKRKKKIEESNEQPQVEVCPKSLVDSVIPIVAQKE 260
Query: 126 --TSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPE 175
+ T++S PT KKKKK+K N+ V + + T P++ G +P+
Sbjct: 261 NAETTTENSNPTP----KKKKKKK----NKNVQDAEQLLTCDPSIAIGAVPD 392
Score = 31.2 bits (69), Expect = 1.4
Identities = 28/103 (27%), Positives = 40/103 (38%)
Frame = +3
Query: 139 SVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLA 198
S KKK+KRK + E E V P ++ V+P KE T P
Sbjct: 135 SEKKKRKRKRKKKIEESNEQPQVEVCPKSLVDSVIPIVAQKENAETTTENSNP------- 293
Query: 199 IPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPIDPEKNKG 241
P+K +K+ N +AE C +P + + D K G
Sbjct: 294 ---TPKKKKKKKNKNVQDAEQLLTC-DPSIAIGAVPDVTKEGG 410
Score = 29.3 bits (64), Expect = 5.1
Identities = 20/75 (26%), Positives = 36/75 (47%), Gaps = 7/75 (9%)
Frame = +3
Query: 204 EKNRKEAEPNESNAEHPRA--CPEPPLGLAIPIDPEKNKG----ENLTPTDENELDRRNR 257
++ RK + E + E P+ CP+ + IPI +K EN PT + + ++N+
Sbjct: 147 KRKRKRKKKIEESNEQPQVEVCPKSLVDSVIPIVAQKENAETTTENSNPTPKKKKKKKNK 326
Query: 258 R-EELVALVTSDEKI 271
++ L+T D I
Sbjct: 327 NVQDAEQLLTCDPSI 371
>CF921716
Length = 367
Score = 44.7 bits (104), Expect = 1e-04
Identities = 36/141 (25%), Positives = 51/141 (35%)
Frame = +1
Query: 106 KKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEP 165
KKKK+ + P + +P K+ + P KKKKK++G + + E
Sbjct: 10 KKKKKKIFTPPRKKKPTKKKNAKKKKGGPPHGKKKKKKKKKRGEKKKKRRGE-------- 165
Query: 166 PAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPE 225
K+ E + P PP G +K+ P E P P+
Sbjct: 166 -------------KQNPPREKKKTAPHPPRG---------KKKKKPPRGGGGEDP---PK 270
Query: 226 PPLGLAIPIDPEKNKGENLTP 246
PP G A P P KN G P
Sbjct: 271 PPGGGAPPRKPPKNNGVKKNP 333
Score = 35.8 bits (81), Expect = 0.055
Identities = 35/126 (27%), Positives = 49/126 (38%), Gaps = 4/126 (3%)
Frame = +2
Query: 106 KKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELAT----V 161
KKKK+ L P + P+ K+T P KKKKK+ G + + + T
Sbjct: 11 KKKKKKFLPPHEKKNPQKKKTQKKKRGGPPTEKKKKKKKKKGGKKKKKEGGKNKTPPGKK 190
Query: 162 CTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPR 221
PP +G E K K P + P+ P G P PE +K ++ E P
Sbjct: 191 KRRPPTPRG----EKKKKNPPGGGGGKT-PQNPRGGGPP--PENPQKTMG*KKTPEEEPG 349
Query: 222 ACPEPP 227
PP
Sbjct: 350 GGAIPP 367
>TC207437 similar to UP|O81922 (O81922) Proline-rich protein, partial (65%)
Length = 1179
Score = 43.1 bits (100), Expect = 3e-04
Identities = 40/131 (30%), Positives = 58/131 (43%), Gaps = 1/131 (0%)
Frame = +3
Query: 107 KKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPP 166
K E V P+P+P P+P++ + + Q K K+ K E + EPP
Sbjct: 279 KSIEIVEPPKPKPPPQPEKPKEPEKPKEPQ----KPKEPEKPKEPEKP--------KEPP 422
Query: 167 AVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEP 226
+ PE KPKEP E P+ PE P P +PEK ++ +P E P P
Sbjct: 423 KPKE---PE-KPKEP---EKPKE-PEKPKEPEKPKEPEKPKEPEKPKEPEKPKPECPPPV 578
Query: 227 PLGLA-IPIDP 236
P+ A +P+ P
Sbjct: 579 PVCPAPVPVCP 611
Score = 38.1 bits (87), Expect = 0.011
Identities = 46/159 (28%), Positives = 62/159 (38%), Gaps = 5/159 (3%)
Frame = +3
Query: 81 QGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPE-PKETSNTDHSEPTQLCS 139
+G I++ + P +P K K+ E EPQ EPE PKE +P +
Sbjct: 261 KGCGTIKSIEIVEPPKPKPPPQPEKPKEPEKPKEPQKPKEPEKPKEP-----EKPKEPPK 425
Query: 140 VKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAI 199
K+ +K K E + EP KPKEP E P+ PE P
Sbjct: 426 PKEPEKPKEPEKPK----------EP----------EKPKEP---EKPKE-PEKPKEPEK 533
Query: 200 PIDPEKNRKEAEPNESNAEHP-RAC---PEPPLGLAIPI 234
P +PEK + E P P C P PP + +PI
Sbjct: 534 PKEPEKPKPECPPPVPVCPAPVPVCPPQPTPPPPVVVPI 650
Score = 32.0 bits (71), Expect = 0.79
Identities = 30/107 (28%), Positives = 43/107 (40%), Gaps = 2/107 (1%)
Frame = +3
Query: 19 EKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLP-EPVVVATAIPITLET 77
E P+P P P+ ++ + K K +K K E+ E + + P EP E
Sbjct: 297 EPPKPKPPPQPEKPKEPEKPKEPQKPKEPEKPKEPEKPKEPPKPKEP-----------EK 443
Query: 78 PKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPE-PEPEP 123
PKE P + P K +K KE +P+PE P P P
Sbjct: 444 PKEPEKPKEPEKPKEPEKPKEPEKPKEPEKPKEPE-KPKPECPPPVP 581
Score = 31.2 bits (69), Expect = 1.4
Identities = 29/115 (25%), Positives = 45/115 (38%)
Frame = +3
Query: 20 KPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPVVVATAIPITLETPK 79
KP+P P PE+ +E K+ ++ K+ K + EP E PK
Sbjct: 306 KPKPPPQPEKPKEPEKPKEPQKPKEPEKPK-------------EP-----------EKPK 413
Query: 80 EQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEP 134
E +P E P + P K +K KE +PE EP++ + +P
Sbjct: 414 EP-PKPKE-----PEKPKEPEKPKEPEKPKEPEKPKEPEKPKEPEKPKEPEKPKP 560
Score = 28.5 bits (62), Expect = 8.8
Identities = 37/132 (28%), Positives = 44/132 (33%), Gaps = 4/132 (3%)
Frame = +3
Query: 342 PDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHIDQAIPSDSNPAVPTNPEH-K 400
P + PE K +EP K K PE + P P P PE K
Sbjct: 321 PQPEKPKEPEKPKEPQKPKEPEK--PKEPEK---------PKEPPKPKEPEKPKEPEKPK 467
Query: 401 TPKKWKKRAKKKNVLL-SEGGKSNEHNGQPTETPVQKSVSPINAPSI--DPTLPTDQPVP 457
P+K K+ K K E K E E P V P P PT P VP
Sbjct: 468 EPEKPKEPEKPKEPEKPKEPEKPKEPEKPKPECPPPVPVCPAPVPVCPPQPTPPPPVVVP 647
Query: 458 IDLATPKHSEGK 469
I + EG+
Sbjct: 648 IGVCCVPCYEGR 683
>TC230046 similar to UP|Q9LU79 (Q9LU79) Gb|AAF21150.1, partial (28%)
Length = 1798
Score = 41.6 bits (96), Expect = 0.001
Identities = 44/195 (22%), Positives = 76/195 (38%), Gaps = 17/195 (8%)
Frame = +3
Query: 17 REEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEG-------IESHEHDQVLLPEPVVVAT 69
+ P P P P +S RR +K++ E ++ HE + P P
Sbjct: 783 KSTSPPPPPPPPPPPPESARHNTRRTHRKVERERESEITVELDDHEFTTIRSPPPAPPTP 962
Query: 70 AIPITLETPKEQGAEPIEATVVHPVP---DQRPAKTHRKKKKKETVLEPQPEPEPEPKET 126
P +++T E+ ++ ++TV + + +KKKK++T +P+ +
Sbjct: 963 PQPPSVKTRSERKSDRRKSTVKREMAMIWASVLSNQRKKKKKQKTKNNENQDPQYDDNAD 1142
Query: 127 SNTDHS-------EPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPK 179
T+++ P L SV RKG ++ ++ +V PP P P K
Sbjct: 1143ELTNNTTVPPPPPPPPPLPSVFHSLFRKGLGKSK---KIHSVSAPPPPP-----PPPPSK 1298
Query: 180 EPCSTEHPRVCPEPP 194
T P PEPP
Sbjct: 1299RKSQTPPP---PEPP 1334
>TC204653 similar to UP|Q9AT20 (Q9AT20) Histone H1 (Fragment), partial (68%)
Length = 1501
Score = 39.7 bits (91), Expect = 0.004
Identities = 39/157 (24%), Positives = 58/157 (36%), Gaps = 4/157 (2%)
Frame = +1
Query: 94 VPDQRPAKTHRK---KKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAE 150
+P RP+ + KKKK +PQP+P+P+PK + K K + A+
Sbjct: 466 LPPTRPSASSSSSPAKKKKPAPAKPQPKPKPKPKP------KPAAKAKDTKATKSKAPAK 627
Query: 151 SNEAVAELATVCTEPPAVQGGVLPEPKPK-EPCSTEHPRVCPEPPLGLAIPIDPEKNRKE 209
A A V +P P PKPK + P+ P G P+ K
Sbjct: 628 PKAAAKPKAKVPAKPK-------PAPKPKAAAAAASKPKPAAAKPKGTKPAAKPKP--KP 780
Query: 210 AEPNESNAEHPRACPEPPLGLAIPIDPEKNKGENLTP 246
A ++ A P A P+ A + G+ P
Sbjct: 781 AAKTKAAAAKPAAKPKAKPAKASRTSTRTSPGKKAAP 891
Score = 35.4 bits (80), Expect = 0.072
Identities = 31/130 (23%), Positives = 48/130 (36%), Gaps = 3/130 (2%)
Frame = +1
Query: 3 SHSQSELLHSKKRRREEKPEPLPSPEQDQEQSC---SKKKRRKKKKIKEEGIESHEHDQV 59
S S S KK+ KP+P P P+ + + K + K K + +
Sbjct: 484 SASSSSSPAKKKKPAPAKPQPKPKPKPKPKPAAKAKDTKATKSKAPAKPKAAAKPKAKVP 663
Query: 60 LLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEP 119
P+P A PK A+P + T P +PA K K +P +P
Sbjct: 664 AKPKPAPKPKAAAAAASKPKPAAAKP-KGTKPAAKPKPKPA-----AKTKAAAAKPAAKP 825
Query: 120 EPEPKETSNT 129
+ +P + S T
Sbjct: 826 KAKPAKASRT 855
>TC204652 similar to UP|H1_PEA (P08283) Histone H1, partial (81%)
Length = 1347
Score = 39.3 bits (90), Expect = 0.005
Identities = 36/141 (25%), Positives = 59/141 (41%), Gaps = 10/141 (7%)
Frame = +3
Query: 94 VPDQRPAKTHRK-----KKKKETVLEPQPEPEPEP----KETSNTDHSEPTQLCSVKKKK 144
+P RPA + KKKK + +P+P+P+P K+T T P + K K
Sbjct: 468 LPPTRPAASSSSSSSPAKKKKPAAAKSKPKPKPKPAAKAKDTKATKSKAPAKPKPAAKPK 647
Query: 145 KRKGAESNEAVAELATVCTEP-PAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDP 203
+ A++ A A+ +P PA +PK +P + P + A+P P
Sbjct: 648 AKAPAKAKPAAKPKASAAAKPKPAA-----AKPKAAKPAAKSKPAAKTK-----AVPAKP 797
Query: 204 EKNRKEAEPNESNAEHPRACP 224
K A+P +++ R P
Sbjct: 798 AAKPK-AKPVKASRTSTRTSP 857
Score = 30.4 bits (67), Expect = 2.3
Identities = 37/170 (21%), Positives = 59/170 (33%), Gaps = 3/170 (1%)
Frame = +3
Query: 3 SHSQSELLHSKKRRREEKPEPLPSP---EQDQEQSCSKKKRRKKKKIKEEGIESHEHDQV 59
S S S K + KP+P P P +D + + SK + K K
Sbjct: 498 SSSSSPAKKKKPAAAKSKPKPKPKPAAKAKDTKATKSKAPAKPKPAAK------------ 641
Query: 60 LLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEP 119
P+ A A P PK A + P + AK+ K K +P +P
Sbjct: 642 --PKAKAPAKAKPAA--KPKASAAAKPKPAAAKPKAAKPAAKSKPAAKTKAVPAKPAAKP 809
Query: 120 EPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQ 169
+ +P + S T S + +K A + + A+ V + V+
Sbjct: 810 KAKPVKASRT---------STRTSPGKKAAPAAKPAAKKVAVAAKKTPVK 932
Score = 29.6 bits (65), Expect = 3.9
Identities = 70/328 (21%), Positives = 97/328 (29%), Gaps = 10/328 (3%)
Frame = +3
Query: 158 LATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNA 217
L ++ TE P V PEP +P E PE K +K EP
Sbjct: 99 LISMATEEPTVAVEPAPEPASADPPPEEKEETKPE-----------AKAKKTKEPKPKKV 245
Query: 218 EHPRACPEPP-----LGLAIPIDPEKNKGENLTPTDENELDRRNRREELVALVTSDEKIV 272
PR P P + AI EK E + L+ K +
Sbjct: 246 SRPRNPPTHPSYEEMVKDAITSLKEKTGSSQYAIAKFIEEKHKQLPPNFKKLLLYHLKKL 425
Query: 273 NPRDHLVPLKSIPGFVLEQQTQPGPVIPVCPNSASLIGQSQTIPVVDTEQKINKRSRMSK 332
LV +K F L P P +S+S + P + K +K
Sbjct: 426 VAAGKLVKVKG--SFKLP------PTRPAASSSSSSSPAKKKKPAAAKSKPKPKPKPAAK 581
Query: 333 MKRKRMNSMPDKHNTEPPETKKRVLDREEPNKCNAKLPETLMLASNCHIDQAIPSDSNPA 392
K + +P K + P K A+ A P + PA
Sbjct: 582 AKDTKATKSKAPAKPKPAAKPK----AKAPAKAKPAAKPKASAAAKPKPAAAKPKAAKPA 749
Query: 393 VPTNPEHKT---PKKWKKRAKKKNVLLSEGGKSNEHNGQPTETPVQKSVSPINAPSIDPT 449
+ P KT P K + K K V S T T K +P P+
Sbjct: 750 AKSKPAAKTKAVPAKPAAKPKAKPVKASR---------TSTRTSPGKKAAPAAKPAAKKV 902
Query: 450 LPTDQPVPIDLATPK--HSEGKMSNAQR 475
+ P+ PK S K + A+R
Sbjct: 903 AVAAKKTPVKSVKPKSVKSPAKKATAKR 986
>CF922281
Length = 520
Score = 39.3 bits (90), Expect = 0.005
Identities = 45/158 (28%), Positives = 58/158 (36%), Gaps = 4/158 (2%)
Frame = +3
Query: 95 PDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEA 154
P P +K+KKKET L P P P PK N P +KKKK+ A+ +
Sbjct: 63 PPGGPQIWGKKEKKKETPLPPMGTPPPPPKGGKN---PPPRGKKPWEKKKKKGFAKPGKR 233
Query: 155 VAELATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNE 214
+ P GG P P + + PP G P K +K N
Sbjct: 234 KNQW------DPLEGGGAKRGPPPPLKKTPKLAFPILRPPGGGQTPPWGGK*KKNPPKNP 395
Query: 215 SNAEHPRACPE--PPLGLAIPIDPEKNKGEN--LTPTD 248
P P+ PP G P P+ KG+ PTD
Sbjct: 396 PGGPPPNLDPKGIPPRGGGNPPPPKTQKGDPKLAPPTD 509
>TC215986 similar to GB|AAC06384.1|1732361|MDU80269 translation initiation
factor 2 beta {Malus x domestica;} , partial (79%)
Length = 1008
Score = 38.1 bits (87), Expect = 0.011
Identities = 29/80 (36%), Positives = 38/80 (47%), Gaps = 3/80 (3%)
Frame = +3
Query: 76 ETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEP-EPEPKETSNTDHSE- 133
ETP E E E P T +KKKKK T+++P +P E ++T N SE
Sbjct: 114 ETPNEMKEEVPEIVPFDP--------TKKKKKKKITIVDPADDPVEKLAEKTENLSVSEG 269
Query: 134 -PTQLCSVKKKKKRKGAESN 152
T +KKKKK+ SN
Sbjct: 270 VETAFAGLKKKKKKPVEMSN 329
>TC226007 similar to UP|Q8LC13 (Q8LC13) Remorin, partial (65%)
Length = 951
Score = 38.1 bits (87), Expect = 0.011
Identities = 41/163 (25%), Positives = 62/163 (37%), Gaps = 35/163 (21%)
Frame = +1
Query: 454 QPVPIDLATPKHSEGKMSNAQRNNVLLSEGVKSNEQPTETPV-------QKSVNPIKAPS 506
Q PI L HS+GKM+ Q V +E SN P V +KSV P +PS
Sbjct: 175 QLFPITLWKDIHSKGKMTEEQSKKVAETESFPSNPAPEPVVVPKEDVAEEKSVIPQPSPS 354
Query: 507 ----------IDPTLPTDQPVPID-----------LATPKH-------SEGKMSKAQRRK 538
++ T + PI+ +AT K E + SKA +
Sbjct: 355 PADESKALVIVEKTSEVAEEKPIEGSVNRDAVLARVATEKRLSLIKAWEESEKSKADNKS 534
Query: 539 KNKRALKSAWSESVHHSSDQNAKTPVQRTESLKLVEAKSVKAK 581
K + SAW S +++ + ++ E K + +K K
Sbjct: 535 HKKLSAISAWENSKKAAAEAELRKIEEQLEKKKAEYGEKLKNK 663
>TC229563
Length = 669
Score = 38.1 bits (87), Expect = 0.011
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Frame = +1
Query: 105 KKKKKETVLEPQPEPEPEPKETSNT-DHSEPTQLCSVKKKKKRKGAESNE 153
++ KK+ + PQPEPEPE KE + ++P + ++ KK +G E+ E
Sbjct: 55 RRTKKQAKIVPQPEPEPEKKEEKPAEEETKPEEEKPPEEAKKEEGGENKE 204
Score = 29.6 bits (65), Expect = 3.9
Identities = 27/110 (24%), Positives = 42/110 (37%), Gaps = 12/110 (10%)
Frame = +1
Query: 93 PVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSE-PTQLCSVKKKKKRKGAES 151
P P+ P K K ++ET +PE E P+E + E + + K++ K E+
Sbjct: 85 PQPEPEPEKKEEKPAEEET----KPEEEKPPEEAKKEEGGENKEEKGGEESKEEAKKEEN 252
Query: 152 NEAVAELATVCTE-----------PPAVQGGVLPEPKPKEPCSTEHPRVC 190
N V + E PP V+ P + S E+P C
Sbjct: 253 NVVVVPYNNIDDEGLKRMMYYYQYPPL---SVIERIPPPQLFSDENPNAC 393
>CF922863
Length = 392
Score = 37.7 bits (86), Expect = 0.014
Identities = 40/144 (27%), Positives = 51/144 (34%), Gaps = 13/144 (9%)
Frame = +1
Query: 99 PAKTHRKKKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAEL 158
P K +KKKKK L P P + K N PTQ+ K KKKRK
Sbjct: 25 PGKISQKKKKKGGGL-PSKTPPKKKKAQKN-----PTQVQHFKPKKKRK----------- 153
Query: 159 ATVCTEPPAVQGGVLPEPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRK---------- 208
+PP Q P PK+ E + PP P P + +K
Sbjct: 154 -----KPPRAQKEPKTPPPPKKRPRGEKKKKTLSPPNPKGAPPGPREGKKKPGGKTPPPP 318
Query: 209 ---EAEPNESNAEHPRACPEPPLG 229
E P N + P+ P+ P G
Sbjct: 319 GGPEKTPRAQN-KFPKKIPKTPRG 387
>TC214054 similar to UP|Q75QC2 (Q75QC2) Glutamate-rich protein, partial (68%)
Length = 1306
Score = 37.7 bits (86), Expect = 0.014
Identities = 37/161 (22%), Positives = 64/161 (38%), Gaps = 2/161 (1%)
Frame = +1
Query: 6 QSELLHSKKRRREEKPEPLPSPEQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLPEPV 65
++E K + P+ EQ + + ++ + E E+ + + P
Sbjct: 139 ENETTEVSKTQETTPVTEAPATEQPAAEVPATEQPAAEAPAPESTTEAPKEETTEAPTET 318
Query: 66 VVATAIPITLETPKEQGAEPIEA--TVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEP 123
V T + E PKE P+E V +++PA ++K E E E EP
Sbjct: 319 VEKTTTEVAPEEPKE---VPVETEEVVAKETEEEKPA-----EEKSEEKTEEVKEEAEEP 474
Query: 124 KETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTE 164
KET+ T+ + + +++ K AES E E+ TE
Sbjct: 475 KETTETESAAAAPPATT--EEENKPAESVETPVEVPVEKTE 591
>BM094772
Length = 421
Score = 37.4 bits (85), Expect = 0.019
Identities = 22/49 (44%), Positives = 30/49 (60%), Gaps = 1/49 (2%)
Frame = +1
Query: 390 NPAVPTNPEHKTPKKWKKRAKKKNVLLSEGGKSNEHNG-QPTETPVQKS 437
+PA +PE P KKR KK+N +EG +S +HN + ETPVQK+
Sbjct: 16 DPATTVDPE---PMTKKKRKKKRNSAKNEGLESEKHNADRIAETPVQKT 153
Score = 36.2 bits (82), Expect = 0.042
Identities = 21/51 (41%), Positives = 30/51 (58%)
Frame = +1
Query: 518 PIDLATPKHSEGKMSKAQRRKKNKRALKSAWSESVHHSSDQNAKTPVQRTE 568
P D AT E K +RKK + + K+ ES H++D+ A+TPVQ+TE
Sbjct: 10 PEDPATTVDPEPMTKK--KRKKKRNSAKNEGLESEKHNADRIAETPVQKTE 156
>TC219933 weakly similar to UP|Q9EQF1 (Q9EQF1) GABA-A epsilon subunit splice
variant (Fragment), partial (7%)
Length = 490
Score = 37.4 bits (85), Expect = 0.019
Identities = 42/162 (25%), Positives = 65/162 (39%), Gaps = 5/162 (3%)
Frame = +1
Query: 69 TAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPEPKETSN 128
+++ ++L+ + EP P P+ P EPQP+PE +P+ T
Sbjct: 79 SSVSLSLKP*HSEMPEPESQNGHEPAPEPEP--------------EPQPQPETKPEPTEQ 216
Query: 129 TD-HSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLPEPKPK-EPCSTEH 186
T H EP +E+V EL P P+P+ EP TE
Sbjct: 217 TQPHLEP----------------KSESVPELE--------------PNPQPESEPVPTEQ 306
Query: 187 PRVCPEPPLGL-AIPIDPEKNRKEA--EPNESNAEHPRACPE 225
+ EP G A P+ + +R+E NE+NA +P P+
Sbjct: 307 TQAPLEPKSGSEADPVVNDADRRETTIHSNEANA-NPSPTPK 429
>TC227676
Length = 1104
Score = 37.4 bits (85), Expect = 0.019
Identities = 25/82 (30%), Positives = 32/82 (38%), Gaps = 16/82 (19%)
Frame = +1
Query: 60 LLPEPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKT----------------H 103
LLP P ++ L K Q P E +HP P +P K H
Sbjct: 238 LLPIPSLLQRVTSFNLSLHKHQPTPP-ETQHLHPEPQPQPEKPPELLRTPSLLERLRSFH 414
Query: 104 RKKKKKETVLEPQPEPEPEPKE 125
K EP+PEPEPEP++
Sbjct: 415 NLSLHKHVQPEPEPEPEPEPEK 480
>TC225160 UP|Q948X9 (Q948X9) Beta-conglycinin alpha-subunit, complete
Length = 2152
Score = 37.0 bits (84), Expect = 0.025
Identities = 29/124 (23%), Positives = 49/124 (39%), Gaps = 10/124 (8%)
Frame = +3
Query: 13 KKRRREEKPEPLPSP----------EQDQEQSCSKKKRRKKKKIKEEGIESHEHDQVLLP 62
K+ +E+P P+P P EQ +EQ +K+ ++ +K EE + EH P
Sbjct: 354 KEEDEDEQPRPIPFPRPQPRQEEEHEQREEQEWPRKEEKRGEKGSEEEQDGREH-----P 518
Query: 63 EPVVVATAIPITLETPKEQGAEPIEATVVHPVPDQRPAKTHRKKKKKETVLEPQPEPEPE 122
P P ++ E E P P + RK+++ E + + E E
Sbjct: 519 RP-----------HQPHDEDEEQDERQFPFPRPPHQKESEERKQEEDEDEEQQRESEESE 665
Query: 123 PKET 126
E+
Sbjct: 666 SSES 677
Score = 29.6 bits (65), Expect = 3.9
Identities = 18/68 (26%), Positives = 34/68 (49%), Gaps = 7/68 (10%)
Frame = +3
Query: 93 PVPDQRPAKTHRK-----KKKKETVLEPQPEPEPEPKETSNTDHSEPTQLCSVKKKKKR- 146
P P RP R+ +K+++ +P+P P P P+ +H + + +K++KR
Sbjct: 297 PRPRPRPQHPEREPQQPGEKEEDEDEQPRPIPFPRPQPRQEEEHEQREEQEWPRKEEKRG 476
Query: 147 -KGAESNE 153
KG+E +
Sbjct: 477 EKGSEEEQ 500
>AW317726 weakly similar to PIR|T17104|T171 translation initiation factor
eIF-2 beta chain - apple tree (fragment), partial (28%)
Length = 449
Score = 36.6 bits (83), Expect = 0.032
Identities = 29/89 (32%), Positives = 44/89 (48%), Gaps = 9/89 (10%)
Frame = +3
Query: 102 THRKKKKKETVLEPQPEP-EPEPKETSNTDHSE--PTQLCSVKKKKKRK------GAESN 152
T +KKKKK T+++P EP E ++T N SE T +KKKKK+ ES
Sbjct: 153 TKKKKKKKITIVDPADEPVEKLAEKTENLSVSEGVETAFTGLKKKKKKPVEMGNLNDESG 332
Query: 153 EAVAELATVCTEPPAVQGGVLPEPKPKEP 181
+A+ +L + P G + P+ + P
Sbjct: 333 DAIEDL----DDHPEDDEGEMVVPQARYP 407
>TC207509
Length = 641
Score = 36.6 bits (83), Expect = 0.032
Identities = 34/141 (24%), Positives = 53/141 (37%)
Frame = +3
Query: 115 PQPEPEPEPKETSNTDHSEPTQLCSVKKKKKRKGAESNEAVAELATVCTEPPAVQGGVLP 174
P+P+P PEP+ S S +Q + E V ++ ++ A +GGV P
Sbjct: 246 PEPQPSPEPQSQSLPPSSPDSQRAWTR--------EDIRYVKDVPSIAPVAYASRGGVAP 401
Query: 175 EPKPKEPCSTEHPRVCPEPPLGLAIPIDPEKNRKEAEPNESNAEHPRACPEPPLGLAIPI 234
P + P + E+ EAE NE +A E + + P+
Sbjct: 402 LPDDRAPVED---------------AAESERRMIEAE-NEMRLRMAKAVEEERMKVPFPL 533
Query: 235 DPEKNKGENLTPTDENELDRR 255
+ K E P D NE R+
Sbjct: 534 LIKPKKNEKPPPLDLNEAIRQ 596
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.306 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,666,039
Number of Sequences: 63676
Number of extensions: 465658
Number of successful extensions: 7703
Number of sequences better than 10.0: 242
Number of HSP's better than 10.0 without gapping: 3408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5077
length of query: 586
length of database: 12,639,632
effective HSP length: 102
effective length of query: 484
effective length of database: 6,144,680
effective search space: 2974025120
effective search space used: 2974025120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0148.5


