

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
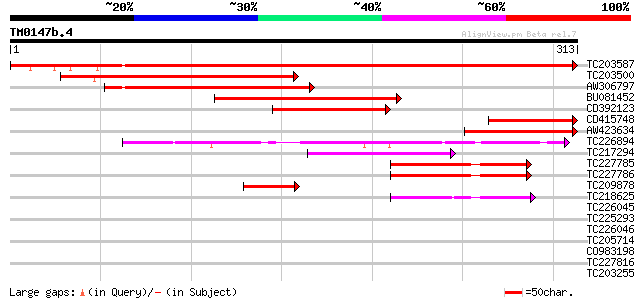
Query= TM0147b.4
(313 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203587 similar to UP|Y230_ARATH (O80934) Protein At2g37660, ch... 483 e-137
TC203500 similar to UP|Y230_ARATH (O80934) Protein At2g37660, ch... 171 4e-43
AW306797 149 1e-36
BU081452 similar to SP|O80934|Y230_ Protein At2g37660 chloropla... 145 2e-35
CD392123 127 8e-30
CD415748 91 7e-19
AW423634 89 3e-18
TC226894 similar to UP|O64701 (O64701) Expressed protein (At2g34... 75 3e-14
TC217294 similar to UP|Q8SKU2 (Q8SKU2) Tic62 protein precursor, ... 65 5e-11
TC227785 55 3e-08
TC227786 55 3e-08
TC209878 51 6e-07
TC218625 homologue to UP|Q8S4X1 (Q8S4X1) UOS1, partial (46%) 48 5e-06
TC226045 similar to UP|Q9FSC6 (Q9FSC6) Cinnamoyl-CoA reductase ... 38 0.007
TC225293 weakly similar to GB|AAM64538.1|21592589|AY086975 cinna... 35 0.034
TC226046 similar to UP|Q7Y0H8 (Q7Y0H8) Cinnamoyl CoA reductase, ... 35 0.057
TC205714 homologue to GB|AAM91372.1|22137054|AY133542 At4g35250/... 33 0.13
CO983198 32 0.49
TC227816 30 1.1
TC203255 UP|Q9ATM4 (Q9ATM4) Plasma membrane integral protein ZmP... 30 1.1
>TC203587 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (76%)
Length = 1832
Score = 483 bits (1242), Expect = e-137
Identities = 258/334 (77%), Positives = 283/334 (84%), Gaps = 21/334 (6%)
Frame = +1
Query: 1 MGMATRVPFV--SATLSPNQCHKYC-------LPSSLKLLT----SLQFSHSSSLSLATH 47
+GMATRVPFV +AT SPN C KYC LP S L+ SL FS S S SL
Sbjct: 88 IGMATRVPFVLCAATTSPNHCLKYCVVAPSLPLPVSSSSLSRHSLSLSFSSSFSSSLGLL 267
Query: 48 --------KRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRT 99
KRV R VVAMA+S +STVLVTGAGGRTG+IVYKKL+ER +QY+ARGLVRT
Sbjct: 268 PLSKKEGVKRVGRRFGVVAMAES-KSTVLVTGAGGRTGQIVYKKLRERPNQYVARGLVRT 444
Query: 100 EESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYF 159
+ESKQ IGA+DDV VGDIRD SI PAIQGIDALIILTSAVP +KPGF+PTKG+RPEFYF
Sbjct: 445 DESKQNIGAADDVIVGDIRDAESIVPAIQGIDALIILTSAVPQIKPGFDPTKGQRPEFYF 624
Query: 160 EDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAE 219
EDGAYPEQVDWIGQKNQID AKAAGVK IVLVGSMGGTDLN+PLNSLGNGNILVWKRKAE
Sbjct: 625 EDGAYPEQVDWIGQKNQIDVAKAAGVKHIVLVGSMGGTDLNHPLNSLGNGNILVWKRKAE 804
Query: 220 QYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFE 279
QYLADSGIPYTIIRAGGLQDK+GG+REL++GKDDE+L+TETRTI+R DVAEVCIQALNFE
Sbjct: 805 QYLADSGIPYTIIRAGGLQDKDGGLRELLVGKDDELLQTETRTISRSDVAEVCIQALNFE 984
Query: 280 EAQFKAFDLASKPEGAGTPTRDFKALFSQITTRF 313
EA+FKAFDLASKPEGAG+ T+DFKALFSQITTRF
Sbjct: 985 EAKFKAFDLASKPEGAGSATKDFKALFSQITTRF 1086
>TC203500 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (34%)
Length = 505
Score = 171 bits (433), Expect = 4e-43
Identities = 92/134 (68%), Positives = 108/134 (79%), Gaps = 3/134 (2%)
Frame = +2
Query: 29 KLLTSLQFSHSSSLSLA---THKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQ 85
+L SL FS ++L+ KRV R SVVAMA+S +STVLVTGAGGRTG+IVYKKL+
Sbjct: 41 QLSLSLSFSLLGLVTLS*KERAKRVGRRFSVVAMAESEKSTVLVTGAGGRTGQIVYKKLK 220
Query: 86 ERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKP 145
ER +QY+ARGLVRT+ESKQ IGA+DDV+VGDIR SI PAIQGIDALIILTSAVP +KP
Sbjct: 221 ERPNQYVARGLVRTDESKQNIGAADDVFVGDIRHAESIVPAIQGIDALIILTSAVPQIKP 400
Query: 146 GFNPTKGERPEFYF 159
GF+PTKG+RP F
Sbjct: 401 GFDPTKGQRPRVLF 442
Score = 30.8 bits (68), Expect = 0.83
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = +3
Query: 151 KGERPEFYFEDGAYPEQ 167
K + EFYF+DGAYPEQ
Sbjct: 417 KDKDQEFYFDDGAYPEQ 467
>AW306797
Length = 346
Score = 149 bits (377), Expect = 1e-36
Identities = 81/116 (69%), Positives = 95/116 (81%)
Frame = +2
Query: 53 RTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDV 112
R VVAMADS ++TVLVTGAGG TG+IVYKKL+ER +QY+ARGLV T+ESKQ IGA+ DV
Sbjct: 2 RFGVVAMADS-KTTVLVTGAGGPTGQIVYKKLRERPNQYVARGLVTTDESKQNIGAAHDV 178
Query: 113 YVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQV 168
V DIRD SIAPAIQG+ ALIILTSAVP +KP F+PT + P+FYFED +Y QV
Sbjct: 179 IVRDIRDADSIAPAIQGLYALIILTSAVPHIKPCFDPTY*QPPKFYFEDVSYSYQV 346
>BU081452 similar to SP|O80934|Y230_ Protein At2g37660 chloroplast
precursor. [Mouse-ear cress] {Arabidopsis thaliana},
partial (20%)
Length = 310
Score = 145 bits (367), Expect = 2e-35
Identities = 74/103 (71%), Positives = 83/103 (79%)
Frame = +2
Query: 114 VGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQ 173
VGDIR SI PAIQGIDALIILTSAVP +KPGF+PTKG+RP FYF+DGAYPEQV
Sbjct: 2 VGDIRHAESIVPAIQGIDALIILTSAVPQIKPGFDPTKGQRPFFYFDDGAYPEQVVCFVP 181
Query: 174 KNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKR 216
IDAAKAAGVK +VL + GTDL++PLN+LGNGNIL WKR
Sbjct: 182 TPPIDAAKAAGVKHLVLKKNKSGTDLSHPLNNLGNGNILGWKR 310
>CD392123
Length = 601
Score = 127 bits (318), Expect = 8e-30
Identities = 59/65 (90%), Positives = 62/65 (94%)
Frame = +1
Query: 146 GFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNS 205
GF+PTKG+RPEFYFE GAYPEQVDWIGQKNQID AKAAGVK IVLVGSMGGTDLN+PLNS
Sbjct: 406 GFDPTKGQRPEFYFEYGAYPEQVDWIGQKNQIDVAKAAGVKHIVLVGSMGGTDLNHPLNS 585
Query: 206 LGNGN 210
LGNGN
Sbjct: 586 LGNGN 600
>CD415748
Length = 633
Score = 90.9 bits (224), Expect = 7e-19
Identities = 44/49 (89%), Positives = 47/49 (95%)
Frame = -1
Query: 265 RPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITTRF 313
R DVAEVCIQALNFEEA+FKAFDLASKPEGAG+ T+DFKALFSQITTRF
Sbjct: 372 RSDVAEVCIQALNFEEAKFKAFDLASKPEGAGSATKDFKALFSQITTRF 226
>AW423634
Length = 187
Score = 89.0 bits (219), Expect = 3e-18
Identities = 45/62 (72%), Positives = 51/62 (81%)
Frame = +2
Query: 252 DDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITT 311
DDE+L+TETRTI D AE+CIQALN EA+F A DLASKP GAG T+D+KALFSQITT
Sbjct: 2 DDELLQTETRTIMTSDDAELCIQALNTCEARFNACDLASKPVGAGAATKDYKALFSQITT 181
Query: 312 RF 313
RF
Sbjct: 182 RF 187
>TC226894 similar to UP|O64701 (O64701) Expressed protein
(At2g34460/T31E10.20), partial (91%)
Length = 1197
Score = 75.5 bits (184), Expect = 3e-14
Identities = 69/258 (26%), Positives = 125/258 (47%), Gaps = 11/258 (4%)
Frame = +2
Query: 63 SRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASD---DVYVGDIRD 119
++ V V GA G TGK + ++L + + G+ +++K T+ +++ + D+ +
Sbjct: 185 AKKKVFVAGATGSTGKRIVEQLLAKGFA-VKAGVRDVDKAKTTLSSANPSLQIVKADVTE 361
Query: 120 -TGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQID 178
+ +A AI ++ + +PG+ D P +VD G N ++
Sbjct: 362 GSDKLAEAIGDDSEAVVCATG---FRPGW-------------DLLAPWKVDNFGTVNLVE 493
Query: 179 AAKAAGVKQIVLVGSM-----GGTDLNNPLNSLGN--GNILVWKRKAEQYLADSGIPYTI 231
A + V + +L+ S+ L NP N G LV K +AE+Y+ SGI YTI
Sbjct: 494 ACRKRNVNRFILISSILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAEKYIRKSGINYTI 673
Query: 232 IRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASK 291
IR GGL++ + +++ +D + + +I+R VAEV ++AL + EA +K ++ S+
Sbjct: 674 IRPGGLRN-DPPTGNVVMEPEDTLYE---GSISRDLVAEVAVEALAYPEAFYKVVEIVSR 841
Query: 292 PEGAGTPTRDFKALFSQI 309
P+ P R + LF I
Sbjct: 842 PD---APKRPYHDLFGSI 886
>TC217294 similar to UP|Q8SKU2 (Q8SKU2) Tic62 protein precursor, partial
(54%)
Length = 1301
Score = 64.7 bits (156), Expect = 5e-11
Identities = 35/83 (42%), Positives = 48/83 (57%), Gaps = 1/83 (1%)
Frame = +2
Query: 165 PEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGN-GNILVWKRKAEQYLA 223
P ++D+ KN IDAA A V +LV S+G + P L +LVWKRKAE+ L
Sbjct: 5 PFRIDYQATKNLIDAATVAKVNHFILVTSLGTNKIGFPAAILNLFWGVLVWKRKAEEALL 184
Query: 224 DSGIPYTIIRAGGLQDKEGGVRE 246
SG+PYTI+R GG++ +E
Sbjct: 185 ASGLPYTIVRPGGMERPTDAFKE 253
>TC227785
Length = 772
Score = 55.5 bits (132), Expect = 3e-08
Identities = 31/78 (39%), Positives = 49/78 (62%)
Frame = +1
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAE 270
+L KR E L SG+ YTI+R G LQ++ GG R LI + + I ++ I+ DVA+
Sbjct: 157 VLKAKRAGEDSLRRSGLGYTIVRPGPLQEEPGGQRALIFDQGNRI----SQGISCADVAD 324
Query: 271 VCIQALNFEEAQFKAFDL 288
+C++AL+ A+ K+FD+
Sbjct: 325 ICVKALHDTTARNKSFDV 378
>TC227786
Length = 988
Score = 55.5 bits (132), Expect = 3e-08
Identities = 31/78 (39%), Positives = 49/78 (62%)
Frame = +1
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAE 270
+L KR E L SG+ YTI+R G LQ++ GG R LI + + I ++ I+ DVA+
Sbjct: 463 VLKAKRAGEDSLRRSGLGYTIVRPGPLQEEPGGQRALIFDQGNRI----SQGISCADVAD 630
Query: 271 VCIQALNFEEAQFKAFDL 288
+C++AL+ A+ K+FD+
Sbjct: 631 ICVKALHDTTARNKSFDV 684
>TC209878
Length = 756
Score = 51.2 bits (121), Expect = 6e-07
Identities = 24/31 (77%), Positives = 27/31 (86%)
Frame = +3
Query: 130 IDALIILTSAVPLMKPGFNPTKGERPEFYFE 160
IDALIILTSAVP +K GF+PTKG R EFYF+
Sbjct: 312 IDALIILTSAVPQIKHGFDPTKGPRSEFYFD 404
>TC218625 homologue to UP|Q8S4X1 (Q8S4X1) UOS1, partial (46%)
Length = 1133
Score = 48.1 bits (113), Expect = 5e-06
Identities = 30/80 (37%), Positives = 45/80 (55%)
Frame = +3
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIARPDVAE 270
IL +K K E L +SGIPY I+R L ++ G +LI + D I T I+R ++A
Sbjct: 513 ILTFKLKGEDLLRESGIPYVIVRPCALTEEPAGA-DLIFDQGDNI----TGKISREEIAL 677
Query: 271 VCIQALNFEEAQFKAFDLAS 290
+C+ AL+ A K F++ S
Sbjct: 678 MCVAALDSPYACDKTFEVKS 737
>TC226045 similar to UP|Q9FSC6 (Q9FSC6) Cinnamoyl-CoA reductase , partial
(93%)
Length = 1254
Score = 37.7 bits (86), Expect = 0.007
Identities = 40/144 (27%), Positives = 59/144 (40%), Gaps = 8/144 (5%)
Frame = +2
Query: 60 ADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEES------KQTIGASD--D 111
A++S T+ VTGAGG + K L E+ Y RG +R + K+ GAS+
Sbjct: 59 AEASSETICVTGAGGFIASWMVKLLLEKG--YTVRGTLRNPDDPKNGHLKEFEGASERLT 232
Query: 112 VYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWI 171
++ D+ S+ I G + S V + PE E
Sbjct: 233 LHKVDLLHLDSVRSVINGCHGVFHTASPVT-----------DNPEEMVEPAV-------S 358
Query: 172 GQKNQIDAAKAAGVKQIVLVGSMG 195
G KN I AA A V+++V S+G
Sbjct: 359 GAKNVIIAAAEAKVRRVVFTSSIG 430
>TC225293 weakly similar to GB|AAM64538.1|21592589|AY086975 cinnamoyl-CoA
reductase-like protein {Arabidopsis thaliana;} , partial
(97%)
Length = 1421
Score = 35.4 bits (80), Expect = 0.034
Identities = 43/156 (27%), Positives = 66/156 (41%), Gaps = 9/156 (5%)
Frame = +1
Query: 48 KRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVR-------TE 100
K+V+ R S+ ++ V VTGA G G V L +R Y V+ T+
Sbjct: 85 KQVKRRASLN*QSNPMSKVVCVTGASGAIGSWVVLLLLQRG--YTVHATVQDIKDENETK 258
Query: 101 ESKQTIGASDDVYVG--DIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFY 158
++ GA ++ D+ D SIA AI+G +I L + + E PE
Sbjct: 259 HLEEMEGAKSHLHFFEMDLLDIDSIAAAIKGCSGVIHLACPNIIGQV-------EDPEKQ 417
Query: 159 FEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSM 194
+ A G N + AAK AGV+++V S+
Sbjct: 418 ILEPAIK------GTVNVLKAAKEAGVERVVATSSI 507
>TC226046 similar to UP|Q7Y0H8 (Q7Y0H8) Cinnamoyl CoA reductase, partial
(58%)
Length = 780
Score = 34.7 bits (78), Expect = 0.057
Identities = 45/164 (27%), Positives = 65/164 (39%), Gaps = 8/164 (4%)
Frame = +1
Query: 40 SSLSLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRT 99
S LSL K+ + +S + S T+ VTGAGG + K L E+ Y RG +R
Sbjct: 40 SLLSLPKKKKKKMPSSESSTGFSE--TICVTGAGGFIASWMVKLLLEKG--YTVRGTLRN 207
Query: 100 EES------KQTIGASD--DVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTK 151
+ K+ GAS ++ D+ S+ I G + S V
Sbjct: 208 PDDPKNGHLKEFEGASQRLTLHKVDLLHLDSVRSVINGCHGVFHTASPVT---------- 357
Query: 152 GERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMG 195
+ PE E G KN I AA A V+++V S+G
Sbjct: 358 -DNPEEMVEPAVN-------GAKNVIIAAAEAKVRRVVFTSSIG 465
>TC205714 homologue to GB|AAM91372.1|22137054|AY133542 At4g35250/F23E12_190
{Arabidopsis thaliana;} , partial (82%)
Length = 1493
Score = 33.5 bits (75), Expect = 0.13
Identities = 51/212 (24%), Positives = 86/212 (40%), Gaps = 5/212 (2%)
Frame = +1
Query: 65 STVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTI----GASDDVYVGDIRDT 120
+ +LV GA G G+ + ++ + Y R LVR + GA+ V D+
Sbjct: 244 TNILVVGATGTLGRQIVRRALDEG--YDVRCLVRPRPAPADFLRDWGAT--VVNADLSKP 411
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAA 180
+I + GI +I + P E P + VDW G+ I A
Sbjct: 412 ETIPATLVGIHTVIDCATGRP-----------EEPI---------KTVDWEGKVALIQCA 531
Query: 181 KAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDK 240
KA G+++ V S+ D + + ++ K E++L DSG+ + IIR G
Sbjct: 532 KAMGIQKYVFY-SIHNCDKHPEV------PLMEIKFCTEKFLRDSGLNHVIIRLCGFMQG 690
Query: 241 EGGVRELIIGKDDEILKTETRT-IARPDVAEV 271
G + I ++ + T+ T IA D ++
Sbjct: 691 LIGQYAVPILEEKSVWGTDAPTRIAYMDTQDI 786
>CO983198
Length = 736
Score = 31.6 bits (70), Expect = 0.49
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = -2
Query: 208 NGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEI 255
+G++L E L +SGIPY I+R L ++ G LI+ + D I
Sbjct: 657 DGHVLGLYEPGEDLLRESGIPYAIVRPCALTEEPAGA-NLILDQGDNI 517
>TC227816
Length = 1221
Score = 30.4 bits (67), Expect = 1.1
Identities = 26/103 (25%), Positives = 43/103 (41%), Gaps = 2/103 (1%)
Frame = +2
Query: 16 PNQCHKYCLPSSLKLLTSLQFSHSSSLS--LATHKRVRTRTSVVAMADSSRSTVLVTGAG 73
PN + C P S L T ++ S S L L + +V T +A S + L+
Sbjct: 743 PNILVEGCDPYSEDLWTEIKISRFSFLGVKLCSRCKVPTINQETGIAGSEPTETLMKT-- 916
Query: 74 GRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGD 116
R+GK++ + ++ Y + LV G+ + VGD
Sbjct: 917 -RSGKVIRPNAKNKNKVYFGQNLVWNWMDSSVKGSGKTIKVGD 1042
>TC203255 UP|Q9ATM4 (Q9ATM4) Plasma membrane integral protein ZmPIP2-7,
complete
Length = 1395
Score = 30.4 bits (67), Expect = 1.1
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = +3
Query: 123 IAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYF-EDGAYPEQ-VDWIGQKNQIDAA 180
+AP G ++ + +P+ G NP + P F D A+ +Q + W+G + AA
Sbjct: 732 LAPLPIGFAVFMVHLATIPVTGTGINPARSFGPAVIFNNDKAWDDQWIYWVGP--FVGAA 905
Query: 181 KAAGVKQIVLVGS 193
AA Q +L GS
Sbjct: 906 VAAIYHQYILRGS 944
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,046,511
Number of Sequences: 63676
Number of extensions: 130154
Number of successful extensions: 944
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 928
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 935
length of query: 313
length of database: 12,639,632
effective HSP length: 97
effective length of query: 216
effective length of database: 6,463,060
effective search space: 1396020960
effective search space used: 1396020960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0147b.4


