

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0145.4
(72 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
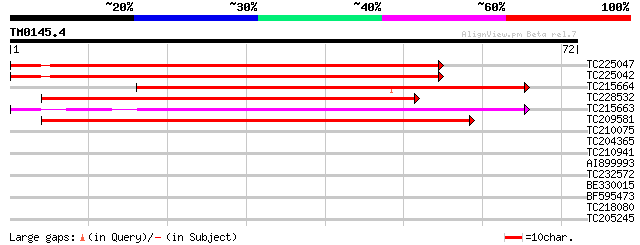
Score E
Sequences producing significant alignments: (bits) Value
TC225047 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-... 94 1e-20
TC225042 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-... 94 1e-20
TC215664 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-... 62 7e-11
TC228532 similar to UP|Q8GZE8 (Q8GZE8) Dehydration-responsive el... 56 4e-09
TC215663 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-... 55 7e-09
TC209581 similar to UP|Q9LKK0 (Q9LKK0) Apetala2 domain-containin... 53 3e-08
TC210075 homologue to PIR|H96667|H96667 AP2-containing DNA-bindi... 33 0.028
TC204365 UP|GLC1_SOYBN (P04776) Glycinin G1 precursor [Contains:... 30 0.30
TC210941 similar to UP|Q9LSN5 (Q9LSN5) Gb|AAD22658.1 (AT3g17120/... 29 0.52
AI899993 27 2.6
TC232572 weakly similar to UP|HDA1_HUMAN (Q13547) Histone deacet... 26 3.4
BE330015 25 5.7
BF595473 homologue to GP|15988117|pdb Chain A Crystal Structure... 25 5.7
TC218080 UP|GLC3_SOYBN (P11828) Glycinin G3 precursor [Contains:... 25 9.8
TC205245 weakly similar to UP|Q754G3 (Q754G3) AFR107Wp, partial ... 25 9.8
>TC225047 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-binding factor
2, partial (42%)
Length = 1886
Score = 94.0 bits (232), Expect = 1e-20
Identities = 46/55 (83%), Positives = 49/55 (88%)
Frame = +3
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK 55
PNLR HQGSSVGGDFGEYKPLHS+VDAKLQAICEGLAE+QKQGKTEK K + K
Sbjct: 1080 PNLR-HQGSSVGGDFGEYKPLHSAVDAKLQAICEGLAELQKQGKTEKPPRKSRSK 1241
>TC225042 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-binding factor
2, partial (40%)
Length = 1705
Score = 94.0 bits (232), Expect = 1e-20
Identities = 46/55 (83%), Positives = 49/55 (88%)
Frame = +3
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK 55
PNLR HQGSSVGGDFGEYKPLHS+VDAKLQAICEGLAE+QKQGKTEK K + K
Sbjct: 1071 PNLR-HQGSSVGGDFGEYKPLHSAVDAKLQAICEGLAELQKQGKTEKPPRKTRSK 1232
>TC215664 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-binding
factor 2, partial (34%)
Length = 1286
Score = 61.6 bits (148), Expect = 7e-11
Identities = 32/52 (61%), Positives = 37/52 (70%), Gaps = 2/52 (3%)
Frame = +2
Query: 17 EYKPLHSSVDAKLQAICEGLAEMQKQGKTEK--KSEKPQKKSKATASKVAPQ 66
EYKP+HS+VDAKL AIC LAEMQKQGKTEK +S K K+ +K PQ
Sbjct: 641 EYKPVHSAVDAKLDAICANLAEMQKQGKTEKGARSAKKSKQGPNQEAKPEPQ 796
>TC228532 similar to UP|Q8GZE8 (Q8GZE8) Dehydration-responsive element
binding protein 3, partial (55%)
Length = 1565
Score = 55.8 bits (133), Expect = 4e-09
Identities = 26/48 (54%), Positives = 35/48 (72%)
Frame = +1
Query: 5 RHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKP 52
RH G+ V G+FG+YKPL SSVD+KLQAICE LA+ +++ + KP
Sbjct: 811 RHHGAFVFGEFGDYKPLPSSVDSKLQAICESLAKQEEKPCCSVEDVKP 954
>TC215663 similar to UP|Q6IVL3 (Q6IVL3) Transcription factor DRE-binding factor
2, partial (33%)
Length = 1777
Score = 55.1 bits (131), Expect = 7e-09
Identities = 31/66 (46%), Positives = 40/66 (59%)
Frame = +2
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATA 60
PNL+ GS G EYKP+ ++VDAKL AIC LAEMQKQGK EK + +K +
Sbjct: 1052 PNLK---GSCPGE---EYKPMQAAVDAKLDAICANLAEMQKQGKNEKGARSGKKSKQGPN 1213
Query: 61 SKVAPQ 66
+ P+
Sbjct: 1214 LEAKPE 1231
>TC209581 similar to UP|Q9LKK0 (Q9LKK0) Apetala2 domain-containing protein,
partial (44%)
Length = 1113
Score = 53.1 bits (126), Expect = 3e-08
Identities = 26/55 (47%), Positives = 35/55 (63%)
Frame = +1
Query: 5 RHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKAT 59
RH G+ V G+FG YKPL S+VDAKLQAIC+ L ++ KT++ K K +
Sbjct: 397 RHHGARVYGEFGNYKPLPSAVDAKLQAICQSLGTKLQKLKTDQNPVVDAKVKKTS 561
>TC210075 homologue to PIR|H96667|H96667 AP2-containing DNA-binding protein,
51686-52693 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (40%)
Length = 1024
Score = 33.1 bits (74), Expect = 0.028
Identities = 18/37 (48%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Frame = +2
Query: 13 GDFGEYKPLHSSVDAKLQAICEGL-AEMQKQGKTEKK 48
GD L SSVDAK+QAIC+ + E K+ T+KK
Sbjct: 437 GDSARLNALKSSVDAKIQAICQKVKKERAKKNATKKK 547
>TC204365 UP|GLC1_SOYBN (P04776) Glycinin G1 precursor [Contains: Glycinin A1A
subunit; Glycinin BX subunit], complete
Length = 1752
Score = 29.6 bits (65), Expect = 0.30
Identities = 14/35 (40%), Positives = 18/35 (51%)
Frame = -3
Query: 36 LAEMQKQGKTEKKSEKPQKKSKATASKVAPQEGCC 70
L + QK G T KK KPQ S+ + +GCC
Sbjct: 1582 LPKRQKWGSTYKKGSKPQLSSETPEVEPGT*KGCC 1478
>TC210941 similar to UP|Q9LSN5 (Q9LSN5) Gb|AAD22658.1 (AT3g17120/K14A17_24),
partial (15%)
Length = 659
Score = 28.9 bits (63), Expect = 0.52
Identities = 15/28 (53%), Positives = 16/28 (56%)
Frame = +3
Query: 44 KTEKKSEKPQKKSKATASKVAPQEGCC* 71
KTE + PQK KAT S P GCC*
Sbjct: 159 KTED*ALYPQKAPKATPSSQRPITGCC* 242
>AI899993
Length = 411
Score = 26.6 bits (57), Expect = 2.6
Identities = 19/66 (28%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Frame = +3
Query: 2 NLRRHQGSSVG--GDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKAT 59
NL HQ +G G+ + + + +AK + E +AE EKK EKP+++ K
Sbjct: 174 NLAFHQSPFIGXMGEEAKQEQVQPVAEAKPEEKKEEMAE-------EKKEEKPEEEKKKE 332
Query: 60 ASKVAP 65
+P
Sbjct: 333 PKPPSP 350
>TC232572 weakly similar to UP|HDA1_HUMAN (Q13547) Histone deacetylase 1
(HD1), partial (5%)
Length = 674
Score = 26.2 bits (56), Expect = 3.4
Identities = 19/62 (30%), Positives = 30/62 (47%), Gaps = 6/62 (9%)
Frame = +1
Query: 2 NLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTE------KKSEKPQKK 55
NL HQ +G GE + ++Q + E E +K+ K E KK EKP++K
Sbjct: 247 NLTFHQSRFIG-QMGE-----EAKQEQVQPVAEAKPEEKKEEKAEEKPAEEKKEEKPEEK 408
Query: 56 SK 57
++
Sbjct: 409 TE 414
>BE330015
Length = 479
Score = 25.4 bits (54), Expect = 5.7
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +2
Query: 5 RHQGSSVGGDFGEYK 19
RHQG++ GG GE+K
Sbjct: 5 RHQGTNAGGGSGEFK 49
>BF595473 homologue to GP|15988117|pdb Chain A Crystal Structure Of Soybean
Proglycinin A1ab1b Homotrimer, partial (7%)
Length = 412
Score = 25.4 bits (54), Expect = 5.7
Identities = 13/35 (37%), Positives = 17/35 (48%)
Frame = +1
Query: 36 LAEMQKQGKTEKKSEKPQKKSKATASKVAPQEGCC 70
L + QK G T KK KPQ S+ + +G C
Sbjct: 262 LPKRQKWGSTYKKGSKPQLSSETPEVEPGT*KGXC 366
>TC218080 UP|GLC3_SOYBN (P11828) Glycinin G3 precursor [Contains: Glycinin A
subunit; Glycinin B subunit], complete
Length = 1708
Score = 24.6 bits (52), Expect = 9.8
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = -1
Query: 40 QKQGKTEKKSEKPQKKSKATASKVAPQEGCC 70
+K G T KK KPQ S+ + +GCC
Sbjct: 1522 RKCGSTCKKGSKPQLSSETP*VEPGS*KGCC 1430
>TC205245 weakly similar to UP|Q754G3 (Q754G3) AFR107Wp, partial (11%)
Length = 372
Score = 24.6 bits (52), Expect = 9.8
Identities = 16/59 (27%), Positives = 27/59 (45%)
Frame = +1
Query: 4 RRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASK 62
R H+ SS D Y S ++ ++ + E + K KKS++P+K + SK
Sbjct: 196 RHHRHSS---DSDSYSSSSLSDYSRSESSSDSEHETSHRSKRHKKSDRPKKNKEKDRSK 363
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,740,267
Number of Sequences: 63676
Number of extensions: 22686
Number of successful extensions: 175
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 175
length of query: 72
length of database: 12,639,632
effective HSP length: 48
effective length of query: 24
effective length of database: 9,583,184
effective search space: 229996416
effective search space used: 229996416
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0145.4


