

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
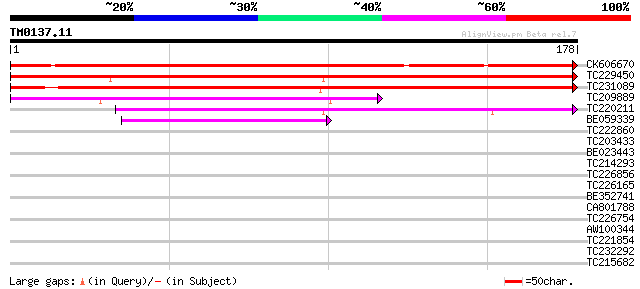
Query= TM0137.11
(178 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CK606670 186 5e-48
TC229450 weakly similar to UP|Q8LJU6 (Q8LJU6) Invertase inhibito... 178 1e-45
TC231089 weakly similar to UP|O82001 (O82001) Tomato invertase i... 161 2e-40
TC209889 100 5e-22
TC220211 85 2e-17
BE059339 43 7e-05
TC222860 weakly similar to PIR|C86366|C86366 protein F26F24.2 [i... 30 0.46
TC203433 similar to PIR|T09390|T09390 21K protein precursor - al... 30 0.60
BE023443 28 1.8
TC214293 weakly similar to UP|Q7XAK4 (Q7XAK4) GID2 protein, part... 27 3.9
TC226856 similar to UP|Q6QTF1 (Q6QTF1) EARLY FLOWERING 5, partia... 27 3.9
TC226165 GAI1 [Glycine max] 27 3.9
BE352741 27 3.9
CA801788 27 5.1
TC226754 similar to UP|Q9XIN0 (Q9XIN0) Expressed protein (BHLH t... 27 5.1
AW100344 similar to PIR|T09390|T093 21K protein precursor - alfa... 27 6.7
TC221854 27 6.7
TC232292 similar to UP|O80872 (O80872) Expressed protein (At2g30... 26 8.7
TC215682 similar to UP|LEU1_SOYBN (Q39891) Probable 2-isopropylm... 26 8.7
>CK606670
Length = 641
Score = 186 bits (472), Expect = 5e-48
Identities = 103/178 (57%), Positives = 126/178 (69%)
Frame = +2
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPR 60
M NLK LS+IC+I +VVA ISMS HCRVLQP+D +L+ +TCK TP P+ CLQ L+ DPR
Sbjct: 86 MLNLKHLSIICSI-LVVAIISMSACHCRVLQPNDLKLIEETCKRTPKPNLCLQLLKGDPR 262
Query: 61 SSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQAT 120
+ SAD LALI+VDVI+AK N I QLLK GG+K AL+ C Y IL DIPQAT
Sbjct: 263 APSADTASLALILVDVIKAKANEAEKTIKQLLKQGGNKKALSECAVDYKGILILDIPQAT 442
Query: 121 QALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
+A++ G+PKFA+D V+D VEA+ CE+ F +GKSPLT GM VARAI R LL
Sbjct: 443 RAVR-GDPKFADDAVSDCAVEADICENRF-NGKSPLTHVKYGMRDVANVARAINRTLL 610
>TC229450 weakly similar to UP|Q8LJU6 (Q8LJU6) Invertase inhibitor-like
protein, partial (24%)
Length = 656
Score = 178 bits (452), Expect = 1e-45
Identities = 92/182 (50%), Positives = 129/182 (70%), Gaps = 4/182 (2%)
Frame = +1
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVL-QPSDAQLVAKTCKNTPYPSACLQFLQADP 59
M+ ++SL+LI +V+ATIS+ + R++ Q ++A L+ +TCK TP+ C+Q+L +DP
Sbjct: 67 MKIMESLALIFYSTLVLATISVPATNSRIIHQKNNANLIEETCKQTPHHDLCIQYLSSDP 246
Query: 60 RSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYNAILKADI 116
RS+ ADVTGLALIMV+VI+ K N L+KI QLL+ + K L+SC RY AI++AD+
Sbjct: 247 RSTEADVTGLALIMVNVIKIKANNALDKIHQLLQKNPEPSQKEPLSSCAARYKAIVEADV 426
Query: 117 PQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRN 176
QA +L+ G+PKFAEDG DA +EA TCE+ FS+GKSPLT+ NN MH + AI+R
Sbjct: 427 AQAVASLQKGDPKFAEDGANDAAIEATTCENSFSAGKSPLTNHNNAMHDVATITAAIVRQ 606
Query: 177 LL 178
LL
Sbjct: 607 LL 612
>TC231089 weakly similar to UP|O82001 (O82001) Tomato invertase inhibitor
precursor, partial (25%)
Length = 788
Score = 161 bits (407), Expect = 2e-40
Identities = 88/182 (48%), Positives = 115/182 (62%), Gaps = 4/182 (2%)
Frame = +2
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPR 60
M NLK L L+ ++ IS+ HCR L P + +L+ TC+ TP + CL+ L+A P
Sbjct: 17 MTNLKPLILLA----IIXMISIPSSHCRTLLPENEKLIENTCRKTPNYNVCLESLKASPG 184
Query: 61 SSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGG----DKPALNSCQGRYNAILKADI 116
SSSADVTGLA IMV ++AK N L +I +L + G + AL+SC +Y +L AD+
Sbjct: 185 SSSADVTGLAQIMVKEMKAKANYALKRIQELQRVGAGPNKQRRALSSCVDKYKTVLIADV 364
Query: 117 PQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRN 176
PQAT+AL+ G+PKFAEDG DA EA CE+ FS+G SPLT +NN MH V AI R
Sbjct: 365 PQATEALQKGDPKFAEDGANDAANEATFCEADFSAGNSPLTKQNNAMHDVAAVTAAIGRL 544
Query: 177 LL 178
LL
Sbjct: 545 LL 550
>TC209889
Length = 427
Score = 100 bits (248), Expect = 5e-22
Identities = 56/122 (45%), Positives = 74/122 (59%), Gaps = 5/122 (4%)
Frame = +3
Query: 1 MENLKSLSLICNIFIVVATISMSIGHC-RVLQPSDAQLVAKTCKNTPYPSACLQFLQADP 59
M NLK L L + +V IS+ HC R L P + +L+ TCK TP + CL+ L+A P
Sbjct: 60 MTNLKPLILFFYLLAIVVMISIPSSHCSRTLLPENEKLIENTCKKTPNYNVCLESLKASP 239
Query: 60 RSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKP----ALNSCQGRYNAILKAD 115
SSSADVTGLA IMV ++AK N L +I +L + G P AL+SC +Y A+L AD
Sbjct: 240 GSSSADVTGLAQIMVKEMKAKANDALKRIQELQRVGASGPNQRRALSSCADKYKAVLIAD 419
Query: 116 IP 117
+P
Sbjct: 420 VP 425
>TC220211
Length = 639
Score = 84.7 bits (208), Expect = 2e-17
Identities = 53/149 (35%), Positives = 77/149 (51%), Gaps = 4/149 (2%)
Frame = +3
Query: 34 DAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLK 93
D LV + CK TP+ C L ++P + +D G+ALIMV+ I L+ I +L+K
Sbjct: 18 DGDLVDQICKKTPFYDLCSSILHSNPLAPKSDPKGMALIMVNGILTNATDTLSYIEELIK 197
Query: 94 GGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFS 150
D + L C Y ++K +PQA A+ G FA + DA E N C+ FS
Sbjct: 198 QTSDEQLEQQLAFCAESYIPVVKYILPQAADAINQGRFGFASYCIVDAQKEVNACDKKFS 377
Query: 151 -SGKSPLTSENNGMHIATEVARAIIRNLL 178
S ++PL+ N+ M +VA AII+ LL
Sbjct: 378 GSTQAPLSDRNDIMQKLVDVAAAIIKLLL 464
>BE059339
Length = 429
Score = 43.1 bits (100), Expect = 7e-05
Identities = 20/66 (30%), Positives = 34/66 (51%)
Frame = +2
Query: 36 QLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGG 95
+L+ CKN C+Q L +DP S AD+ LALI + + +G+LN +++
Sbjct: 32 ELIKSICKNRGNDELCMQVLSSDPDSDHADLQELALISLKAAASNASGILNDCKRMIDNQ 211
Query: 96 GDKPAL 101
+P +
Sbjct: 212 DLEPKI 229
>TC222860 weakly similar to PIR|C86366|C86366 protein F26F24.2 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(8%)
Length = 616
Score = 30.4 bits (67), Expect = 0.46
Identities = 31/139 (22%), Positives = 51/139 (36%), Gaps = 7/139 (5%)
Frame = +2
Query: 24 IGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVT-------GLALIMVDV 76
IG+C + +C TPYPS C +++ S+ D + L + MV
Sbjct: 29 IGYCLLYSLLLVHGKQLSCNETPYPSVCKHYIETTKTLSALDASPSSFHDMALKVTMVQA 208
Query: 77 IQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVA 136
++A V N K K A C Y L Q +++ + N +
Sbjct: 209 MEA-YKLVSNMDLNNFKDKRAKSAWEDCLELYENTLY----QLKRSMNSNNLNDRMTWQS 373
Query: 137 DAGVEANTCESGFSSGKSP 155
+ TC++GF+ P
Sbjct: 374 ASIANHQTCQNGFTDFNLP 430
>TC203433 similar to PIR|T09390|T09390 21K protein precursor - alfalfa
{Medicago sativa;} , partial (88%)
Length = 825
Score = 30.0 bits (66), Expect = 0.60
Identities = 31/160 (19%), Positives = 59/160 (36%), Gaps = 16/160 (10%)
Frame = +1
Query: 7 LSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADV 66
++ I N ++ + +S H + + +C T YP+ C+Q L + D
Sbjct: 37 MTTISNCSLLFLFLFLSTLHIASTLSTPTNFIKSSCSTTQYPALCIQSLSVYASTIQQDP 216
Query: 67 TGLALIMVDVIQAKTNGVLNKISQLLKGGGDKP----ALNSCQGRYNAILKADIPQATQA 122
L + + T +++ K G KP AL C + + + +++
Sbjct: 217 HELVQTALSLSLNHTEATKTFVAKCNKFRGLKPREYAALKDCAEE----ISDSVDRLSRS 384
Query: 123 LK------------TGNPKFAEDGVADAGVEANTCESGFS 150
LK T + E V+ A + +TC GF+
Sbjct: 385 LKELKLCKVKGEDFTWHISNVETWVSSALTDESTCGDGFA 504
>BE023443
Length = 375
Score = 28.5 bits (62), Expect = 1.8
Identities = 19/74 (25%), Positives = 28/74 (37%)
Frame = +1
Query: 32 PSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQL 91
PS + + CK T YP C L A RSS +D G + + +
Sbjct: 103 PSSSSSSSVACKGTLYPKLCRSILSA-IRSSPSDPYGYGKFSIKQSLKQARKLAKVFEDF 279
Query: 92 LKGGGDKPALNSCQ 105
L+ P+LN +
Sbjct: 280 LQRHQKSPSLNHAE 321
>TC214293 weakly similar to UP|Q7XAK4 (Q7XAK4) GID2 protein, partial (34%)
Length = 1212
Score = 27.3 bits (59), Expect = 3.9
Identities = 19/70 (27%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Frame = -2
Query: 9 LICNIFIVVATISMS----IGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSA 64
+ + F+ +++ S+S + HC P+ A + A TC NT + L + P +SS+
Sbjct: 686 VFAHCFVQISSQSLSSCAVLCHCLFTHPAIASVRASTCFNT--SNRRLSSKRPSPSTSSS 513
Query: 65 DVTGLALIMV 74
LA ++V
Sbjct: 512 FSIALAFLLV 483
>TC226856 similar to UP|Q6QTF1 (Q6QTF1) EARLY FLOWERING 5, partial (24%)
Length = 1247
Score = 27.3 bits (59), Expect = 3.9
Identities = 18/54 (33%), Positives = 24/54 (44%)
Frame = -2
Query: 14 FIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVT 67
FIV A + H + QPS A + K Y S+ L AD R + +D T
Sbjct: 862 FIVHALTIIQHRHSQRSQPSSAPRAFMSSKKAVYESSMLCVFGADDRLTDSDFT 701
>TC226165 GAI1 [Glycine max]
Length = 1313
Score = 27.3 bits (59), Expect = 3.9
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = +3
Query: 139 GVEANTCESGFSSGKSPLTSENNGM 163
G A T G+SSGKS L E GM
Sbjct: 120 GSNAGTSSCGYSSGKSNLWEEEGGM 194
>BE352741
Length = 558
Score = 27.3 bits (59), Expect = 3.9
Identities = 22/100 (22%), Positives = 41/100 (41%)
Frame = +1
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPR 60
+ K S +C + + + + + V P+ + CK+TP PS C L P+
Sbjct: 19 LSQTKMASKLCFLILFLILLPLFASSETVPNPTTSVSPGTACKSTPDPSYCKSVL--PPQ 192
Query: 61 SSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPA 100
+ + G + + QA+ LN + + L+ G A
Sbjct: 193 NGNVYDYGRFSVKKSLSQARK--FLNLVDKYLQRGSSLSA 306
>CA801788
Length = 457
Score = 26.9 bits (58), Expect = 5.1
Identities = 20/84 (23%), Positives = 33/84 (38%), Gaps = 5/84 (5%)
Frame = +3
Query: 92 LKGGGDKP-----ALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCE 146
L+GG + P A+ A++ ALK P +ED + V +
Sbjct: 135 LEGGSNSPLAAAAAIAKIGAEATAVISQVQASVINALKKAFPVGSEDQIVKHQVNGVNED 314
Query: 147 SGFSSGKSPLTSENNGMHIATEVA 170
GFS ++PL + + EV+
Sbjct: 315 FGFSESENPLILSDQDSPVNAEVS 386
>TC226754 similar to UP|Q9XIN0 (Q9XIN0) Expressed protein (BHLH transcription
factor), partial (11%)
Length = 761
Score = 26.9 bits (58), Expect = 5.1
Identities = 20/67 (29%), Positives = 31/67 (45%)
Frame = +1
Query: 62 SSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQATQ 121
++ DVT + + M ++ + Q +KGGG A +S N ++ PQATQ
Sbjct: 136 ANRDVTRMEIFM---------SLVRLLDQTVKGGG---ASSSNAIDNNMMVYQSFPQATQ 279
Query: 122 ALKTGNP 128
TG P
Sbjct: 280 ITATGRP 300
>AW100344 similar to PIR|T09390|T093 21K protein precursor - alfalfa,
partial (26%)
Length = 364
Score = 26.6 bits (57), Expect = 6.7
Identities = 17/73 (23%), Positives = 30/73 (40%), Gaps = 7/73 (9%)
Frame = +2
Query: 13 IFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFL-------QADPRSSSAD 65
+F+ ++T+ H + + +C T YP+ C+Q L Q DP
Sbjct: 2 LFLFLSTL-----HIASTLSTPTNFIKSSCSTTQYPALCIQSLSVYASTIQQDPHELVQT 166
Query: 66 VTGLALIMVDVIQ 78
V L+L + I+
Sbjct: 167VLSLSLNHTEAIK 205
>TC221854
Length = 667
Score = 26.6 bits (57), Expect = 6.7
Identities = 12/37 (32%), Positives = 18/37 (48%)
Frame = +2
Query: 96 GDKPALNSCQGRYNAILKADIPQATQALKTGNPKFAE 132
G P+ G+ + L A +PQ + GNP FA+
Sbjct: 221 GFNPSRQLEYGQNDMYLNAQVPQPNHQFQQGNPPFAQ 331
>TC232292 similar to UP|O80872 (O80872) Expressed protein
(At2g30010/F23F1.7), partial (6%)
Length = 690
Score = 26.2 bits (56), Expect = 8.7
Identities = 13/41 (31%), Positives = 18/41 (43%)
Frame = -1
Query: 32 PSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALI 72
P Q + T YPS C FL + +S TGL ++
Sbjct: 474 PGRHQWLQSTAITPLYPSGCASFLYCESVVTSKKYTGLVIL 352
>TC215682 similar to UP|LEU1_SOYBN (Q39891) Probable 2-isopropylmalate
synthase (Alpha-isopropylmalate synthase) (Alpha-IPM
synthetase) (Late nodulin 56) (N-56) , partial (8%)
Length = 1481
Score = 26.2 bits (56), Expect = 8.7
Identities = 15/43 (34%), Positives = 23/43 (52%), Gaps = 3/43 (6%)
Frame = +2
Query: 30 LQPSDAQLVAKTCKNTPYPSAC-LQFLQADPRSS--SADVTGL 69
LQP D ++ TC+NT Y C + ++ + P + A TGL
Sbjct: 68 LQPFDFMMMFSTCENTIYAHLCNVGYI*SSPNPTVMGASCTGL 196
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,256,534
Number of Sequences: 63676
Number of extensions: 86382
Number of successful extensions: 420
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 412
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 412
length of query: 178
length of database: 12,639,632
effective HSP length: 91
effective length of query: 87
effective length of database: 6,845,116
effective search space: 595525092
effective search space used: 595525092
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0137.11


