

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.1
(630 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
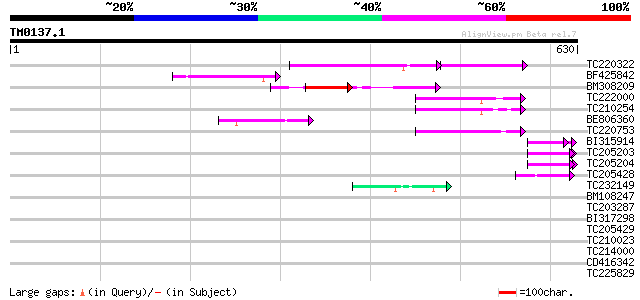
Score E
Sequences producing significant alignments: (bits) Value
TC220322 120 1e-42
BF425842 83 3e-16
BM308209 74 3e-13
TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transp... 73 3e-13
TC210254 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F1... 73 4e-13
BE806360 similar to GP|6175165|gb| Mutator-like transposase {Ara... 59 7e-09
TC220753 51 1e-06
BI315914 similar to PIR|T09854|T098 proline-rich cell wall prote... 45 1e-04
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 45 1e-04
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 45 1e-04
TC205428 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 43 5e-04
TC232149 42 8e-04
BM108247 weakly similar to PIR|H84710|H847 Mutator-like transpos... 42 0.001
TC203287 weakly similar to UP|Q41645 (Q41645) Extensin (Fragment... 41 0.001
BI317298 similar to GP|5139695|dbj| expressed in cucumber hypoco... 40 0.002
TC205429 weakly similar to UP|O22600 (O22600) Glycine-rich prote... 40 0.002
TC210023 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 40 0.002
TC214000 40 0.003
CD416342 similar to SP|P42698|D111_ DNA-damage-repair/toleration... 40 0.003
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 40 0.004
>TC220322
Length = 1264
Score = 120 bits (302), Expect(2) = 1e-42
Identities = 71/171 (41%), Positives = 94/171 (54%), Gaps = 3/171 (1%)
Frame = +2
Query: 312 QAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFN 371
Q AK+T V FE M LK +N +A ++L PK++W K F+ PK D + NN E FN
Sbjct: 2 QCAKSTTVAEFEGHMAHLKTINCQAWEYLNKWPKQAWTKAHFSTIPKVDNICNNTCEVFN 181
Query: 372 ATILLARDKPILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLAWETKSSNNWVP 431
IL R KPI+TM++ IRSY+M AA K P KRL E +N W P
Sbjct: 182 FRILQYRCKPIITMLEEIRSYIMRTMAARKVKLSGKPGPLCLVQYKRLEKEFHFANQWTP 361
Query: 432 QACGH---AKYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGIPCRHAVAAIS 479
CG +YEV + G++ V+L +CTC W L G+PCRHA+A I+
Sbjct: 362 IWCGDNMGLRYEVH--MWGNKVEVNLAGWTCTCGVWQLTGMPCRHAIATIT 508
Score = 71.6 bits (174), Expect(2) = 1e-42
Identities = 37/99 (37%), Positives = 49/99 (49%), Gaps = 2/99 (2%)
Frame = +1
Query: 479 SFNGGHAESYIDSYYGRDAYQATYAHGITPINGKRMWPYTPDAPILPPLYKRKPGRPKKL 538
S GG E + +AY TY H I P+ G + W T +PP + + GRPKK
Sbjct: 508 SHKGGKPEDMCHEWLSIEAYNKTYQHFIEPVQGPQYWAQTQYTHPVPPYKRVQRGRPKKN 687
Query: 539 RRRDPHEDDT--DEGSRPNSRHRCGRCGTIGHNIRTCTN 575
R+R ED+ + R + CGRCG HNIR+C N
Sbjct: 688 RKRSVDEDNVTGHKLKRKLAEFTCGRCGQTNHNIRSCKN 804
>BF425842
Length = 414
Score = 83.2 bits (204), Expect = 3e-16
Identities = 47/125 (37%), Positives = 65/125 (51%), Gaps = 5/125 (4%)
Frame = +3
Query: 181 NRYYICLEGCKRGFVAGCRPFIGLDGCHLKTQYGGILLCAVGRDPNDQYFPIAFAVVESE 240
NR + C GF C+P + +DG L +Y LL A+G+D + FP+AFA+VESE
Sbjct: 27 NRVF*AFNPCIEGFKY-CKPLVQVDGTFLTGKYNDTLLTAIGQDGSRDNFPLAFAIVESE 203
Query: 241 CKESWKWFLELLLADIGGLRRWIFISDQQKGLLSVFDELM-----PGVEHRFCLRHLYAN 295
KE+W WFL L + ISD+ GLL+ P V +C+RH+ +N
Sbjct: 204 TKEAWIWFLHYL*RYVTPQPNLCIISDRGTGLLATLQSKHVGWNGPDVSSVYCIRHIASN 383
Query: 296 FKKKF 300
F K F
Sbjct: 384 FNK*F 398
>BM308209
Length = 435
Score = 73.6 bits (179), Expect = 3e-13
Identities = 33/52 (63%), Positives = 40/52 (76%)
Frame = +2
Query: 329 LKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSEAFNATILLARDK 380
+KEVN EA+DWL+ K W KH F+ Y KCDVLMNNL +AFN+TIL AR+K
Sbjct: 122 IKEVNNEAYDWLIKVGTKLWRKHTFSYYTKCDVLMNNLCDAFNSTILCAREK 277
Score = 59.7 bits (143), Expect = 4e-09
Identities = 54/191 (28%), Positives = 77/191 (40%), Gaps = 2/191 (1%)
Frame = +3
Query: 290 RHLYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWC 349
RH+Y NFK KF GG ++R+LM MR K EEA D M K+
Sbjct: 6 RHIYNNFKNKFVGGTLLRNLM---------------MRAAKASFEEAWDEKMNISKR*IM 140
Query: 350 KHAFTLYPKCDVLMNNLSEAFNATILLARDKPI--LTMIDWIRSYLMGRFAALNEKFQRY 407
K P L L+ + L+ + +T + + + ++ L +
Sbjct: 141 K------PMTG*LRLELNYGVSTHFLITLNVMY**ITCV----TLSIAQYCVLGK----- 275
Query: 408 NHPFMPKPLKRLAWETKSSNNWVPQACGHAKYEVKSVLDGDQFIVDLRQRSCTCNWWDLV 467
S NW GH +EV + D+FIV+L SC+ N W++
Sbjct: 276 ------------------SGNWFATLSGHLVFEVSHSMLSDKFIVNLNSWSCS*NLWEIT 401
Query: 468 GIPCRHAVAAI 478
GI CRHA AAI
Sbjct: 402 GILCRHACAAI 434
>TC222000 weakly similar to PIR|H84710|H84710 Mutator-like transposase
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (12%)
Length = 617
Score = 73.2 bits (178), Expect = 3e-13
Identities = 43/130 (33%), Positives = 60/130 (46%), Gaps = 7/130 (5%)
Frame = +2
Query: 451 IVDLRQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPIN 510
IV++ SC+C W L GIPC HA AA+ ++ + +++ TYA I PI
Sbjct: 17 IVNIGSHSCSCRDWQLNGIPCSHAAAALISCRKDVYAFSQKCFTAASFRDTYAETIHPIP 196
Query: 511 GKRMWPYTPDAP-------ILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRC 563
GK W T ++ + PP +R PGRP+K D+ +R C RC
Sbjct: 197 GKLEWSKTGNSSMDDNILVVRPPKLRRPPGRPEKRMC-------VDDLNREKHTVHCSRC 355
Query: 564 GTIGHNIRTC 573
GH RTC
Sbjct: 356 NQTGHYKRTC 385
>TC210254 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17A9_9
{Arabidopsis thaliana;} , partial (6%)
Length = 814
Score = 72.8 bits (177), Expect = 4e-13
Identities = 41/127 (32%), Positives = 59/127 (46%), Gaps = 4/127 (3%)
Frame = +1
Query: 451 IVDLRQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPIN 510
IVD+ C+C W L G+PC HA+A G Y Y+ + Y+ TYA I P+
Sbjct: 64 IVDIDNWDCSCKGWQLTGVPCCHAIAVFECVGRSPYDYCSRYFTVENYRLTYAESIHPVP 243
Query: 511 GKRMWPYTPDAP----ILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTI 566
P ++ ++PP KR PGRPK + D R + +C +C +
Sbjct: 244 NVDKPPVQGESTALVMVIPPPTKRPPGRPKM-----KQVESIDIIKR---QLQCSKCKGL 399
Query: 567 GHNIRTC 573
GHN +TC
Sbjct: 400 GHNRKTC 420
>BE806360 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (17%)
Length = 431
Score = 58.9 bits (141), Expect = 7e-09
Identities = 33/109 (30%), Positives = 58/109 (52%), Gaps = 4/109 (3%)
Frame = +2
Query: 233 AFAVVESECKESWKWFLE----LLLADIGGLRRWIFISDQQKGLLSVFDELMPGVEHRFC 288
AF VV+ E ++W WFL LL + R +SD+QKG++ + P H FC
Sbjct: 2 AFGVVDEENDDNWMWFLSELHNLLEIHTENMLRLTILSDRQKGIVDGVEASFPTAFHGFC 181
Query: 289 LRHLYANFKKKFGGGVVIRDLMMQAAKATYVHLFEEKMRELKEVNEEAH 337
++HL +F+K+F +++ +L+ +AA A +++ LK ++ H
Sbjct: 182 MQHLSDSFRKEFNNTMLV-NLLWEAANALTELNLKQRFWRLKRYHKMLH 325
>TC220753
Length = 1312
Score = 51.2 bits (121), Expect = 1e-06
Identities = 32/124 (25%), Positives = 51/124 (40%), Gaps = 2/124 (1%)
Frame = +2
Query: 452 VDLRQRSCTCNWWDLVGIPCRHAVAAISFNGGHAESYIDSYYGRDAYQATYAHGITPING 511
V L + C + + +PC HA+AA +F + + ++D Y + Y H +
Sbjct: 215 VKLNEWLCDYGQFQALRLPCSHAIAACAFCNLNYDDFVDPVYKLENIFKVYQHHFHSLGS 394
Query: 512 KRMWPYTPDAPILPPLYKRK--PGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHN 569
+ WP + KR+ GRP R + ++ S PN +C C T GHN
Sbjct: 395 EDTWPQYLGPHFMSDPSKRRQISGRPATTRIHNEMDE-----SIPNRLKKCSLCKTEGHN 559
Query: 570 IRTC 573
C
Sbjct: 560 QSNC 571
>BI315914 similar to PIR|T09854|T098 proline-rich cell wall protein - upland
cotton, partial (37%)
Length = 246
Score = 45.1 bits (105), Expect = 1e-04
Identities = 18/45 (40%), Positives = 23/45 (51%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
PP+S P P + P A+ PP P A+ PP P +T PP P
Sbjct: 55 PPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTPPPSSP 189
Score = 43.5 bits (101), Expect = 3e-04
Identities = 19/54 (35%), Positives = 24/54 (44%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPVV 629
PP+S P P + P A+ PP P + PP P + PP P A P V
Sbjct: 85 PPSSPPPASPPPSSPPPASPPPSSPPPSTPPPSSPPPFSPPPATPPPATPPPAV 246
Score = 40.8 bits (94), Expect = 0.002
Identities = 17/52 (32%), Positives = 24/52 (45%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP+S P P P ++ PP P ++ PP P ++ PP P A P
Sbjct: 70 PPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTPPPSSPPPFSPPPATPPP 225
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/54 (33%), Positives = 25/54 (45%)
Frame = +1
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
+ PP S P P + P ++ PP P A+ PP P A+ PP P + P
Sbjct: 22 STPPPS-PLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTPPP 180
Score = 38.1 bits (87), Expect = 0.012
Identities = 15/53 (28%), Positives = 26/53 (48%)
Frame = +1
Query: 575 NPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
+P +S P P + P ++ PP P ++ PP P ++ PP P ++ P
Sbjct: 37 SPLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTPPPSSPPP 195
Score = 36.2 bits (82), Expect = 0.045
Identities = 16/52 (30%), Positives = 25/52 (47%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP S P P + P ++ PP P ++ PP P ++ PP P ++ P
Sbjct: 13 PPASTPPPS-PLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPP 165
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 45.1 bits (105), Expect = 1e-04
Identities = 20/52 (38%), Positives = 25/52 (47%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP+S P P + P A+ PP P ++ PP P AT PP P A P
Sbjct: 450 PPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPP 605
Score = 44.3 bits (103), Expect = 2e-04
Identities = 19/52 (36%), Positives = 25/52 (47%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP+S P P + P A+ PP P A+ PP P ++ PP P A P
Sbjct: 420 PPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPP 575
Score = 42.4 bits (98), Expect = 6e-04
Identities = 18/54 (33%), Positives = 24/54 (44%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPVV 629
PP+S P P P ++ PP P ++ PP P + PP P A P V
Sbjct: 435 PPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAV 596
Score = 40.0 bits (92), Expect = 0.003
Identities = 18/54 (33%), Positives = 26/54 (47%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
+ PP S P P + P ++ PP P A+ PP P A+ PP P ++ P
Sbjct: 387 STPPPS-PLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPP 545
Score = 37.4 bits (85), Expect = 0.020
Identities = 19/56 (33%), Positives = 27/56 (47%), Gaps = 2/56 (3%)
Frame = +3
Query: 574 TNPPTSQ--PTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
++PP SQ P P P ++ PP P ++ PP P A+ PP P A+ P
Sbjct: 351 SSPPPSQSPPPASTPPPSPL-SSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPP 515
Score = 36.2 bits (82), Expect = 0.045
Identities = 16/52 (30%), Positives = 25/52 (47%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP S P P + P ++ PP P ++ PP P ++ PP P ++ P
Sbjct: 378 PPASTPPPS-PLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPP 530
Score = 34.3 bits (77), Expect = 0.17
Identities = 16/51 (31%), Positives = 23/51 (44%)
Frame = +3
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
P P+ P P A+ PP P ++ PP P ++ PP P A+ P
Sbjct: 336 PPPVPSSPPPSQSPPPASTPPPSPL-SSPPPSSPPPSSPPPSSPPPASPPP 485
Score = 34.3 bits (77), Expect = 0.17
Identities = 16/45 (35%), Positives = 17/45 (37%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
PP+S P P P A PP P A PP T PP
Sbjct: 510 PPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPP 644
Score = 34.3 bits (77), Expect = 0.17
Identities = 20/57 (35%), Positives = 22/57 (38%), Gaps = 2/57 (3%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQ--PPTVAAQNPVVS 630
PP S P P + P PP P A PP P A PP P +P VS
Sbjct: 495 PPASPPPSSPPPSSP-----PPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPVS 650
Score = 32.3 bits (72), Expect = 0.65
Identities = 14/51 (27%), Positives = 21/51 (40%)
Frame = +3
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
P+S P P T P P ++ PP P ++ PP P ++ P
Sbjct: 348 PSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPPSSPPP 500
Score = 32.3 bits (72), Expect = 0.65
Identities = 18/53 (33%), Positives = 23/53 (42%), Gaps = 4/53 (7%)
Frame = +3
Query: 572 TCTNPPTSQ----PTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
T PPT Q P + P P+ + P Q P A+ PP P + P PP
Sbjct: 282 TSPAPPTPQASPPPVQSSPPPVPS-SPPPSQSPPPASTPPPSPLSSPPPSSPP 437
Score = 29.3 bits (64), Expect = 5.5
Identities = 14/45 (31%), Positives = 20/45 (44%), Gaps = 2/45 (4%)
Frame = +3
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQ--PTEATQPPQP 619
P++ P P+ P P P ++ PP Q P +T PP P
Sbjct: 276 PSTSPAPPTPQASPPPVQSSPP-PVPSSPPPSQSPPPASTPPPSP 407
Score = 29.3 bits (64), Expect = 5.5
Identities = 17/53 (32%), Positives = 20/53 (37%), Gaps = 4/53 (7%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQP----TEAAQPPQQPTEATQPPQPPTVAA 624
PP+S P P P A PP P T P P ++ P P V A
Sbjct: 525 PPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPVSSPAPSPSKVFA 683
Score = 28.5 bits (62), Expect = 9.4
Identities = 14/52 (26%), Positives = 20/52 (37%)
Frame = +3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP VP + P + PP T P P ++ PP P ++ P
Sbjct: 318 PPVQSSPPPVPSSPPPSQSPPPAS-TPPPSPLSSPPPSSPPPSSPPPSSPPP 470
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 45.1 bits (105), Expect = 1e-04
Identities = 20/52 (38%), Positives = 24/52 (45%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP S P P P A+ PP P A+ PP P A+ PP P A+ P
Sbjct: 328 PPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATPPPASPPP 483
Score = 43.9 bits (102), Expect = 2e-04
Identities = 22/56 (39%), Positives = 25/56 (44%), Gaps = 1/56 (1%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPP-QPPTVAAQNPVVS 630
PP S P P P A+ PP P A+ PP P AT PP PP P+ S
Sbjct: 388 PPASPPPASPPPASPPPASPPPATPPPASPPPFSPPPATPPPATPPPALTPTPLSS 555
Score = 43.1 bits (100), Expect = 4e-04
Identities = 20/52 (38%), Positives = 23/52 (43%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP S P P P A+ PP P A+ PP P A+ PP P A P
Sbjct: 358 PPASPPPASPPPASPPPASPPPASPPPASPPPATPPPASPPPFSPPPATPPP 513
Score = 42.4 bits (98), Expect = 6e-04
Identities = 19/55 (34%), Positives = 23/55 (41%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPVVS 630
PP S P P P A+ PP P A PP P + PP P A P ++
Sbjct: 373 PPASPPPASPPPASPPPASPPPASPPPATPPPASPPPFSPPPATPPPATPPPALT 537
Score = 41.2 bits (95), Expect = 0.001
Identities = 19/52 (36%), Positives = 24/52 (45%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
P +S P P + P A+ PP P A+ PP P A+ PP P A P
Sbjct: 313 PLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATPPP 468
Score = 41.2 bits (95), Expect = 0.001
Identities = 18/53 (33%), Positives = 25/53 (46%)
Frame = +1
Query: 575 NPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
+PP + P + P A+ PP P A+ PP P A+ PP P A+ P
Sbjct: 280 SPPPASTPPPAPLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPP 438
Score = 39.3 bits (90), Expect = 0.005
Identities = 18/47 (38%), Positives = 23/47 (48%)
Frame = +1
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
++PP + P P P A+ PP P A+ PP P AT PP P
Sbjct: 349 SSPPPASP----PPASPPPASPPPASPPPASPPPASPPPATPPPASP 477
Score = 38.9 bits (89), Expect = 0.007
Identities = 17/51 (33%), Positives = 22/51 (42%)
Frame = +1
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
P P P P ++ PP P A+ PP P A+ PP P A+ P
Sbjct: 301 PPPAPLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPP 453
Score = 36.2 bits (82), Expect = 0.045
Identities = 18/52 (34%), Positives = 21/52 (39%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP S P P P A+ PP P A PP P A P + A +P
Sbjct: 418 PPASPPPASPPPATPPPASPPPFSPPPATPPPATPPPALTPTPLSSPPATSP 573
Score = 35.0 bits (79), Expect = 0.10
Identities = 17/51 (33%), Positives = 21/51 (40%)
Frame = +1
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
P+S P P T P P A+ PP P A+ PP P A+ P
Sbjct: 256 PSSPPPAQSPPPASTPPPAPLSSPPPASPPPSSPPPASPPPASPPPASPPP 408
Score = 34.7 bits (78), Expect = 0.13
Identities = 17/52 (32%), Positives = 23/52 (43%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
P S P P P ++ PP P ++ PP P A+ PP P A+ P
Sbjct: 271 PAQSPPPASTPPPAPL-SSPPPASPPPSSPPPASPPPASPPPASPPPASPPP 423
Score = 32.0 bits (71), Expect = 0.85
Identities = 16/54 (29%), Positives = 22/54 (40%)
Frame = +1
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
++PP Q + + P A PP T P P A+ PP P A+ P
Sbjct: 217 SSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPASPPPSSPPPASPPP 378
Score = 31.6 bits (70), Expect = 1.1
Identities = 18/53 (33%), Positives = 23/53 (42%), Gaps = 4/53 (7%)
Frame = +1
Query: 572 TCTNPPTSQ----PTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPP 620
T PPT Q P + P P+ + P Q P A+ PP P + P PP
Sbjct: 190 TSPAPPTPQSSPPPIQSSPPPVPS-SPPPAQSPPPASTPPPAPLSSPPPASPP 345
Score = 30.8 bits (68), Expect = 1.9
Identities = 19/51 (37%), Positives = 20/51 (38%), Gaps = 2/51 (3%)
Frame = +1
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQP-PQQPTEATQP-PQPPTVAA 624
PP + P P P A PP P A P P AT P P P V A
Sbjct: 448 PPATPPPASPPPFSPPPATPPPATPPPALTPTPLSSPPATSPAPAPAKVVA 600
Score = 28.5 bits (62), Expect = 9.4
Identities = 14/45 (31%), Positives = 21/45 (46%), Gaps = 2/45 (4%)
Frame = +1
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQ--PTEATQPPQP 619
P++ P P++ P P P ++ PP Q P +T PP P
Sbjct: 184 PSTSPAPPTPQSSPPPIQSSPP-PVPSSPPPAQSPPPASTPPPAP 315
>TC205428 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (5%)
Length = 720
Score = 42.7 bits (99), Expect = 5e-04
Identities = 23/65 (35%), Positives = 27/65 (41%)
Frame = -2
Query: 563 CGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTV 622
C + RT PPT PT P PT PP P PP+ P PP+PP +
Sbjct: 419 CSSFQGKGRTPLPPPTPVPTPP-PIPPPTPLPVPPNPPIPPVGPPRPPPSPPPPPKPPPI 243
Query: 623 AAQNP 627
A P
Sbjct: 242 PAPAP 228
>TC232149
Length = 1005
Score = 42.0 bits (97), Expect = 8e-04
Identities = 39/143 (27%), Positives = 54/143 (37%), Gaps = 34/143 (23%)
Frame = -3
Query: 382 ILTMIDWIRSYLMGRFAALNEKFQRYNHPFMPKPLKRLAWETKSSN----NWVPQACGHA 437
IL M + IR+Y+M + + K P +P R+ SN NWV G
Sbjct: 427 ILNMCEDIRTYIMCKMKSDKLKMATRLRPLVPMQQSRIEKGKVESNK*TANWVGDPDG-T 251
Query: 438 KYEVKSVLDGDQFIVDLRQRSCTCNWWDLVGI---------------------------- 469
+++V + VDL+ S TC W L+GI
Sbjct: 250 RFKVCHY--DTRLNVDLQN*SFTCRMWQLIGIPLS*LNLSILDIMYCVTMNLMFFWFIYI 77
Query: 470 --PCRHAVAAISFNGGHAESYID 490
PCRHA+ AI +N E Y D
Sbjct: 76 GMPCRHAIVAIRYNYHIPEGYSD 8
>BM108247 weakly similar to PIR|H84710|H847 Mutator-like transposase
[imported] - Arabidopsis thaliana, partial (4%)
Length = 265
Score = 41.6 bits (96), Expect = 0.001
Identities = 28/78 (35%), Positives = 41/78 (51%)
Frame = +2
Query: 309 LMMQAAKATYVHLFEEKMRELKEVNEEAHDWLMTHPKKSWCKHAFTLYPKCDVLMNNLSE 368
L+ +A+ AT F+EKM E++EV+ EA WL W F + L +N+ E
Sbjct: 23 LLWKASHATTTIAFKEKMGEIEEVSPEAAKWLQQFXPSQWALVHFK-GTRFGHLSSNIEE 199
Query: 369 AFNATILLARDKPILTMI 386
FN IL R+ PI+ +I
Sbjct: 200 -FNKWILDTRELPIIXVI 250
>TC203287 weakly similar to UP|Q41645 (Q41645) Extensin (Fragment), partial
(14%)
Length = 967
Score = 41.2 bits (95), Expect = 0.001
Identities = 24/63 (38%), Positives = 27/63 (42%), Gaps = 6/63 (9%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQ--PTEATQPPQ----PPTVAAQNP 627
T PPT+ P+ P + P A PP P A PP P AT PP P AA P
Sbjct: 141 TQPPTTTPSPPPPRSAPAPAPTPPATPPPATPPPAAATPPPATSPPATTPTPTPTAAPTP 320
Query: 628 VVS 630
S
Sbjct: 321 ASS 329
Score = 36.2 bits (82), Expect = 0.045
Identities = 20/59 (33%), Positives = 26/59 (43%), Gaps = 9/59 (15%)
Frame = +3
Query: 574 TNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEA---------TQPPQPPTVA 623
T+PP + PT P PT A+ P P +A P P A +PP PP+ A
Sbjct: 267 TSPPATTPTP-TPTAAPTPASSPATSPVPSASSPPAPGTAGPAPGPAGGAEPPPPPSAA 440
>BI317298 similar to GP|5139695|dbj| expressed in cucumber hypocotyls
{Cucumis sativus}, partial (34%)
Length = 381
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/57 (36%), Positives = 26/57 (44%), Gaps = 3/57 (5%)
Frame = +1
Query: 577 PTSQPTEDVPETEPTEAAQPPQQPTEAAQP---PQQPTEATQPPQPPTVAAQNPVVS 630
PT+ P P T+P A+ PP P A P P P + PP P VAA V+
Sbjct: 73 PTTTPAAPAPATKPPAASPPPANPPPAPVPVSSPPAPVPVSSPPAPVPVAAPTTFVA 243
>TC205429 weakly similar to UP|O22600 (O22600) Glycine-rich protein, partial
(49%)
Length = 611
Score = 40.4 bits (93), Expect = 0.002
Identities = 23/65 (35%), Positives = 29/65 (44%)
Frame = +3
Query: 563 CGTIGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTV 622
C + RT PPT PT P T P PP P +PP+ P + PP+PP +
Sbjct: 282 CSSFQGKGRTPLPPPTPVPTPP-PPTPPPVPPNPPIPPVVPPRPPRPP--PSPPPKPPPI 452
Query: 623 AAQNP 627
A P
Sbjct: 453 PAPTP 467
>TC210023 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (12%)
Length = 819
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/68 (35%), Positives = 31/68 (45%), Gaps = 2/68 (2%)
Frame = -3
Query: 562 RCGTIGH--NIRTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQP 619
+C +I H +IR+ PP + P P QPPQ P + QPP + PPQP
Sbjct: 502 KCSSIPHLLSIRSPPQPPQPLLSNLFPHPPP----QPPQPPPQPPQPPLFNIDPHPPPQP 335
Query: 620 PTVAAQNP 627
P A P
Sbjct: 334 PPQPAPQP 311
Score = 35.8 bits (81), Expect = 0.059
Identities = 22/56 (39%), Positives = 24/56 (42%), Gaps = 4/56 (7%)
Frame = -3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTE-AAQPPQQP---TEATQPPQPPTVAAQNP 627
PP P P + PPQ P + A QPPQ P PPQPP A Q P
Sbjct: 406 PPQPPPQPPQPPLFNIDPHPPPQPPPQPAPQPPQPPLLIKFPQPPPQPPQPAPQRP 239
Score = 35.4 bits (80), Expect = 0.077
Identities = 19/38 (50%), Positives = 22/38 (57%), Gaps = 3/38 (7%)
Frame = -3
Query: 586 PETEPTEAAQPPQQPT--EAAQPPQQPTE-ATQPPQPP 620
P+ P A QPPQ P + QPP QP + A Q PQPP
Sbjct: 343 PQPPPQPAPQPPQPPLLIKFPQPPPQPPQPAPQRPQPP 230
Score = 34.3 bits (77), Expect = 0.17
Identities = 20/61 (32%), Positives = 24/61 (38%), Gaps = 15/61 (24%)
Frame = -3
Query: 575 NPPTSQPTEDVPETE--------PTEAAQPPQQPTEAAQPP-------QQPTEATQPPQP 619
+PP P + P+ P QPPQ + QPP P A QPPQP
Sbjct: 352 HPPPQPPPQPAPQPPQPPLLIKFPQPPPQPPQPAPQRPQPPLLIKFPQPPPQPAPQPPQP 173
Query: 620 P 620
P
Sbjct: 172 P 170
Score = 33.9 bits (76), Expect = 0.22
Identities = 21/56 (37%), Positives = 25/56 (44%), Gaps = 4/56 (7%)
Frame = -3
Query: 577 PTSQPTEDVPETEPT----EAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNPV 628
P QP + P+ + QPP QP A QPPQ P P PP A Q P+
Sbjct: 277 PPPQPPQPAPQRPQPPLLIKFPQPPPQP--APQPPQPPLLIKFPQPPPQPAPQPPL 116
Score = 32.3 bits (72), Expect = 0.65
Identities = 19/52 (36%), Positives = 19/52 (36%)
Frame = -3
Query: 576 PPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
PP P P PPQ P A Q PQ P P PP A Q P
Sbjct: 334 PPQPAPQPPQPPLLIKFPQPPPQPPQPAPQRPQPPLLIKFPQPPPQPAPQPP 179
Score = 31.6 bits (70), Expect = 1.1
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 6/41 (14%)
Frame = -3
Query: 586 PETEPTEAAQPPQQP--TEAAQPPQQPTE----ATQPPQPP 620
P+ P A QPPQ P + QPP QP +T PQPP
Sbjct: 214 PQPPPQPAPQPPQPPLLIKFPQPPPQPAPQPPLSTAFPQPP 92
>TC214000
Length = 659
Score = 40.0 bits (92), Expect = 0.003
Identities = 27/79 (34%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Frame = +3
Query: 537 KLRRRDPHEDDTDEGSR-PNSRHRCGRCGTIGHNIRTCTNPPT-SQPTEDVPETEPTEAA 594
+LRRR+ ED D R N+ + C RC +GHN RTC N P +P + E E +
Sbjct: 6 RLRRREVDED*NDHKLRGTNTTN*CWRCHKLGHNKRTCKNSPIHPKPAKTRVEGEGAPSE 185
Query: 595 QPPQQPTEAAQPPQQPTEA 613
+ Q TEA
Sbjct: 186 EQQSHVQVGPNGTQVQTEA 242
>CD416342 similar to SP|P42698|D111_ DNA-damage-repair/toleration protein
DRT111 chloroplast precursor. [Mouse-ear cress],
partial (24%)
Length = 624
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/57 (35%), Positives = 31/57 (54%)
Frame = -1
Query: 571 RTCTNPPTSQPTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
R+ + PP+S+P + P PT + PP P+ PP P + T P PP++A+ P
Sbjct: 303 RSASPPPSSRPPK--PSYGPTPSPSPPPSPSSPPPPPPFPRK-TPPSSPPSLASSPP 142
Score = 33.1 bits (74), Expect = 0.38
Identities = 21/66 (31%), Positives = 27/66 (40%), Gaps = 9/66 (13%)
Frame = -1
Query: 571 RTCTNPPTSQPTEDVPETEPTEAAQPPQQ---------PTEAAQPPQQPTEATQPPQPPT 621
+T + PPTS P P P +A PP PT + PP P+ PP PP
Sbjct: 360 KTTSPPPTSGPP--APRWLPPRSASPPPSSRPPKPSYGPTPSPSPPPSPSS---PPPPPP 196
Query: 622 VAAQNP 627
+ P
Sbjct: 195 FPRKTP 178
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 39.7 bits (91), Expect = 0.004
Identities = 30/105 (28%), Positives = 37/105 (34%)
Frame = +2
Query: 521 APILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCTNPPTSQ 580
AP P R P P P T SRP R + + P TS
Sbjct: 323 APPSSPPCSRPPAPPPA-----PPATSTSPTSRP------ARSASPPKTTTAPSTPSTSN 469
Query: 581 PTEDVPETEPTEAAQPPQQPTEAAQPPQQPTEATQPPQPPTVAAQ 625
P+ P T P+ + PP P PP P +T P P AA+
Sbjct: 470 PSRKCPTTSPSSKSAPPLAPPTPRHPPPPPPPST*SPLCPNKAAR 604
Score = 36.2 bits (82), Expect = 0.045
Identities = 31/125 (24%), Positives = 42/125 (32%), Gaps = 5/125 (4%)
Frame = +2
Query: 508 PINGKRMWPYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRC--GRCGT 565
P WP+TP +P P PK+ P S P++ C T
Sbjct: 77 PTTSPACWPHTPASP-PSTTTSPSPTWPKRSTAARP------SPSWPSTMPPCPPSSTST 235
Query: 566 IGHNIRTCTNPPTSQPTEDVPETEPTEAAQPPQQP---TEAAQPPQQPTEATQPPQPPTV 622
++P TS T P++ PP P A PP P +T P P
Sbjct: 236 SPSPPSKTSSPSTSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATSTSPTSRPAR 415
Query: 623 AAQNP 627
+A P
Sbjct: 416 SASPP 430
Score = 33.5 bits (75), Expect = 0.29
Identities = 33/138 (23%), Positives = 43/138 (30%), Gaps = 24/138 (17%)
Frame = +2
Query: 515 WPYTPDAPILPPLYKRKPGRPKKLRRRDPHEDDTDEGSRPNSRHRCGRCGTIGHNIRTCT 574
WP T P P P P K T S P S R T + C+
Sbjct: 191 WPSTMP-PCPPSSTSTSPSPPSKTSSPSTSSSTT---SAPKSSTRST---TAPPSSPPCS 349
Query: 575 NPPTSQP----TEDVPETEPTEAAQPPQ--------------------QPTEAAQPPQQP 610
PP P T P + P +A PP+ P+ + PP P
Sbjct: 350 RPPAPPPAPPATSTSPTSRPARSASPPKTTTAPSTPSTSNPSRKCPTTSPSSKSAPPLAP 529
Query: 611 TEATQPPQPPTVAAQNPV 628
PP PP + +P+
Sbjct: 530 PTPRHPPPPPPPST*SPL 583
Score = 30.8 bits (68), Expect = 1.9
Identities = 18/59 (30%), Positives = 28/59 (46%), Gaps = 6/59 (10%)
Frame = +2
Query: 575 NPPTSQPT--EDVPETEPTEAAQP----PQQPTEAAQPPQQPTEATQPPQPPTVAAQNP 627
+PPT+ P P + P+ P P++ T A P P+ T PP PP+ + +P
Sbjct: 71 SPPTTSPACWPHTPASPPSTTTSPSPTWPKRSTAARPSPSWPS--TMPPCPPSSTSTSP 241
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,905,626
Number of Sequences: 63676
Number of extensions: 590799
Number of successful extensions: 9216
Number of sequences better than 10.0: 428
Number of HSP's better than 10.0 without gapping: 5086
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7442
length of query: 630
length of database: 12,639,632
effective HSP length: 103
effective length of query: 527
effective length of database: 6,081,004
effective search space: 3204689108
effective search space used: 3204689108
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0137.1


