

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
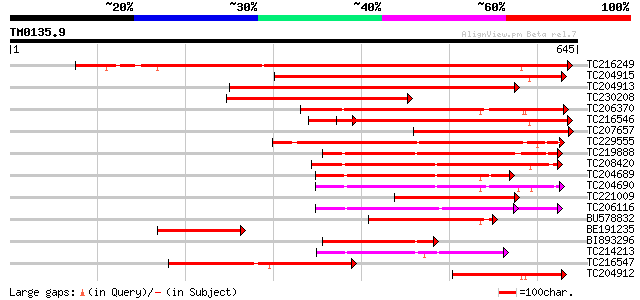
Query= TM0135.9
(645 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216249 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 468 e-132
TC204915 weakly similar to GB|AAH46286.1|28279482|BC046286 spast... 399 e-111
TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 387 e-107
TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 379 e-105
TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, part... 353 2e-97
TC216546 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, ... 329 2e-90
TC207657 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic ... 328 3e-90
TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteaso... 283 2e-76
TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus heli... 222 4e-58
TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%) 205 4e-53
TC204689 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type A... 193 2e-49
TC204690 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type A... 192 3e-49
TC221009 homologue to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, pa... 184 8e-47
TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 h... 170 2e-42
BU578832 170 2e-42
BE191235 163 3e-40
BI893296 similar to GP|21553404|gb 26S proteasome regulatory par... 160 2e-39
TC214213 homologue to UP|PRSA_BRACM (O23894) 26S protease regula... 147 2e-35
TC216547 145 4e-35
TC204912 weakly similar to GB|AAH46286.1|28279482|BC046286 spast... 145 5e-35
>TC216249 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 2222
Score = 468 bits (1203), Expect = e-132
Identities = 264/589 (44%), Positives = 373/589 (62%), Gaps = 23/589 (3%)
Frame = +2
Query: 75 YWINVKNIEHDLDAQSQDCYLAMEALREVLNSKQ---PLIVYFPDGSQWLHKSVPKSIRS 131
++ NV ++ + A + L + +L EV+ S+ P I++ D + + V
Sbjct: 128 FFCNVTDLRLESSAVEELDKLLIHSLFEVVFSESRSAPFILFMKDAEKSI---VGNGDSH 298
Query: 132 EFFHKVEEMFDQLSGPIVLICGQNKAQSGTKEKGQ-----FTMILPNFARLAKL--PLSL 184
F K+E + D V++ G + KEK FT N L L P S
Sbjct: 299 SFKSKLENLPDN-----VVVIGSHTQNDSRKEKSHPGGLLFTKFGSNQTALLDLAFPDSF 463
Query: 185 KRLTDKGQKTSDDDE-INKLFSNVVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELR 243
RL D+G++ + + KLF N ++I P+DE LLA++K+QL+ D + + + NL+ LR
Sbjct: 464 GRLHDRGKEAPKQNRTLTKLFPNKITIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLR 643
Query: 244 KVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESL 303
VL + C L L D LT + AEK++GWA +H+L KL L ES+
Sbjct: 644 TVLGRCGMECEGLETLCIKDQTLTNENAEKIIGWALSHHLMQNS-EAKPDSKLVLSCESI 820
Query: 304 EIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGALEDVKKA 362
I L+ ++ S+ +SLK++ ++EFE ++ V+PP +I V FDDIGALE VK
Sbjct: 821 LYGIGILQSIQNESKSLKKSLKDVVTENEFEKRLLADVIPPSDIDVTFDDIGALEKVKDT 1000
Query: 363 LNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTL 422
L ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AKA+ATEAGANFI++S S++
Sbjct: 1001LKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAIATEAGANFINISMSSI 1180
Query: 423 TSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWD 482
TSKWFG+ EK KA+FS ASK++P +IFVDEVDS+LG R EHEA R+M+NEFM WD
Sbjct: 1181TSKWFGEGEKYVKAVFSLASKISPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWD 1360
Query: 483 GLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPD 542
GLR+KE +R+L+L ATNRPFDLD+AVIRR+PRR+ V+LPDA NR KIL++IL E L PD
Sbjct: 1361GLRTKETERVLVLAATNRPFDLDEAVIRRMPRRLMVNLPDAPNRAKILKVILAKEELSPD 1540
Query: 543 FQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK-----------AGDNNITNVAL 591
D +A++TDGYSGSDLKNLC+TAA+RP++E+LE+EK ++ +
Sbjct: 1541VDLDAVASMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKERAAALAEGQPAPALCSSGDV 1720
Query: 592 RPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
R LN++DF + +V SV+ ++V++ EL +WNE+YGEGG+R K +
Sbjct: 1721RSLNMEDFKYAHQQVCASVSSESVNMTELLQWNELYGEGGSRVKKALSY 1867
>TC204915 weakly similar to GB|AAH46286.1|28279482|BC046286 spastic
paraplegia 4 homolog {Mus musculus;} , partial (21%)
Length = 1470
Score = 399 bits (1026), Expect = e-111
Identities = 196/344 (56%), Positives = 266/344 (76%), Gaps = 12/344 (3%)
Frame = +1
Query: 302 SLEIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGALEDVK 360
SL I+ L+G++ ++ +SLK++ ++EFE ++ V+PP +IGV FDDIGALE+VK
Sbjct: 130 SLNYGINILQGIQNENKNLKKSLKDVVTENEFEKKLLADVIPPTDIGVTFDDIGALENVK 309
Query: 361 KALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGS 420
L ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AKA+ATEAGANFI++S S
Sbjct: 310 DTLKELVMLPLQRPELFCKGQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMS 489
Query: 421 TLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAA 480
++TSKWFG+ EK KA+FS ASK+AP +IFVDEVDS+LG R EHEA R+M+NEFM
Sbjct: 490 SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPSEHEAMRKMKNEFMVN 669
Query: 481 WDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLD 540
WDGLR+K+ +R+L+L ATNRPFDLD+AVIRRLPRR+ V+LPDA NR+KILR+IL E+L
Sbjct: 670 WDGLRTKDKERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNREKILRVILVKEDLA 849
Query: 541 PDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVA---------- 590
PD F+ +AN+TDGYSGSDLKNLC+TAA+ P++E+LE+EK + + +
Sbjct: 850 PDVDFEAIANMTDGYSGSDLKNLCVTAAHCPIREILEKEKKERSLALSESKPLPGLCGSG 1029
Query: 591 -LRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTR 633
+RPL +DDF + +V SV+ ++ ++NEL +WN++YGEGG+R
Sbjct: 1030DIRPLKMDDFRYAHEQVCASVSSESTNMNELLQWNDLYGEGGSR 1161
>TC204913 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 1080
Score = 387 bits (993), Expect = e-107
Identities = 189/331 (57%), Positives = 253/331 (76%), Gaps = 1/331 (0%)
Frame = +3
Query: 251 LSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRL 310
L C DL L D LT + EK++GWA +++ ++ KL + ES++ L
Sbjct: 6 LDCPDLETLCIKDHTLTTESVEKIIGWAISYHFMHSSEASIRDSKLVISAESIKYGHKIL 185
Query: 311 KGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVIL 369
+G++ ++ +SLK++ ++EFE ++ V+PP +IGV FDDIGALE+VK+ L ELV+L
Sbjct: 186 QGIQNENKNMKKSLKDVVTENEFEKKLLTDVIPPTDIGVTFDDIGALENVKETLKELVML 365
Query: 370 PMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGD 429
P++RPELF G L +P KGILLFGPPGTGKT++AKA+ATEAGANFI++S S++TSKWFG+
Sbjct: 366 PLQRPELFGKGQLAKPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGE 545
Query: 430 AEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKEN 489
EK KA+FS ASK+AP +IFVDEVDS+LG R EHEA R+M+NEFM WDGLR+K+
Sbjct: 546 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDK 725
Query: 490 QRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLA 549
+RIL+L ATNRPFDLD+AVIRRLPRR+ V+LPDA NR KI+R+IL E+L PD F+ +A
Sbjct: 726 ERILVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNRGKIVRVILAKEDLAPDVDFEAIA 905
Query: 550 NLTDGYSGSDLKNLCITAAYRPVQELLEEEK 580
N+TDGYSGSDLKNLC+TAA P++++LE+EK
Sbjct: 906 NMTDGYSGSDLKNLCVTAAQCPIRQILEKEK 998
>TC230208 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (45%)
Length = 642
Score = 379 bits (972), Expect = e-105
Identities = 189/212 (89%), Positives = 204/212 (96%)
Frame = +1
Query: 247 EEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIA 306
EEHQLSC+DLL +++D +ILTK KAEKVVGWAKNHYLSSCLLP +KG++L LPRESLEIA
Sbjct: 7 EEHQLSCMDLLLVNTDSIILTKHKAEKVVGWAKNHYLSSCLLPSIKGERLYLPRESLEIA 186
Query: 307 ISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNEL 366
+SRLKG ETMSR+PSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNEL
Sbjct: 187 VSRLKGQETMSRKPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNEL 366
Query: 367 VILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKW 426
VILPMRRPELF+ GNLLRP KGILLFGPPGTGKTL+AKALATEAGANFIS++GSTLTSKW
Sbjct: 367 VILPMRRPELFSRGNLLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKW 546
Query: 427 FGDAEKLTKALFSFASKLAPVIIFVDEVDSLL 458
FGDAEKLTKALFSFASKLAPVI+FVDEVDSLL
Sbjct: 547 FGDAEKLTKALFSFASKLAPVIVFVDEVDSLL 642
>TC206370 similar to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (38%)
Length = 1502
Score = 353 bits (905), Expect = 2e-97
Identities = 183/329 (55%), Positives = 240/329 (72%), Gaps = 24/329 (7%)
Frame = +2
Query: 331 EFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGIL 390
EFE V+P EIGV F DIGAL+++K++L ELV+LP+RRP+LF G LL+P +GIL
Sbjct: 2 EFEKRIRPEVIPANEIGVTFADIGALDEIKESLQELVMLPLRRPDLFK-GGLLKPCRGIL 178
Query: 391 LFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIF 450
LFGPPGTGKT++AKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ A+K+AP IIF
Sbjct: 179 LFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVAPTIIF 358
Query: 451 VDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR 510
VDEVDS+LG R EHEA R+++NEFM WDGL + N++IL+L ATNRPFDLD+A+IR
Sbjct: 359 VDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTGPNEQILVLAATNRPFDLDEAIIR 538
Query: 511 RLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLKNLCITA 567
R RRI V LP +NR+ IL+ +L KHENLD F +LA +T+GY+GSDLKNLCITA
Sbjct: 539 RFERRILVGLPSVENREMILKTLLAKEKHENLD----FKELATMTEGYTGSDLKNLCITA 706
Query: 568 AYRPVQELLEEEKAGD-----------------NN----ITNVALRPLNLDDFVQSKSKV 606
AYRPV+EL+++E+ D NN + LRPLN++D Q+K++V
Sbjct: 707 AYRPVRELIQQERMKDMEKKKREAEGQSSEDASNNKDKEEKEITLRPLNMEDMRQAKTQV 886
Query: 607 GPSVAYDAVSVNELRKWNEMYGEGGTRTK 635
S A + +NEL+ WN++YGEGG+R K
Sbjct: 887 AASFASEGSVMNELKHWNDLYGEGGSRKK 973
>TC216546 weakly similar to UP|SAP1_YEAST (P39955) SAP1 protein, partial
(14%)
Length = 1321
Score = 329 bits (844), Expect = 2e-90
Identities = 159/280 (56%), Positives = 211/280 (74%), Gaps = 11/280 (3%)
Frame = +3
Query: 372 RRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAE 431
R P F GN P KGILLFGPPGTGKT++AKA+ATEAGANFI++S S++TSKWFG+ E
Sbjct: 99 RGPSYFAKGN*PXPCKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGE 278
Query: 432 KLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQR 491
K KA+FS ASK+AP +IFVDEVDS+LG R EHEA R+M+NEFM WDGLR+K+ +R
Sbjct: 279 KYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTER 458
Query: 492 ILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANL 551
+L+L ATNRPFDLD+AVIRRLPRR+ V+LPDA NR KIL++IL+ E+L D D +A++
Sbjct: 459 VLVLAATNRPFDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILEKEDLSSDIDMDAIASM 638
Query: 552 TDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVA-----------LRPLNLDDFV 600
TDGYSGSDLKNLC+TAA+RP++E+LE+EK + +R LN++DF
Sbjct: 639 TDGYSGSDLKNLCVTAAHRPIKEILEKEKKEQAAAVSEGRPAPALSGSGDIRSLNMEDFK 818
Query: 601 QSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
+ +V SV+ +++++ EL++WNE+YGEGG+R K +
Sbjct: 819 YAHQQVCASVSSESINMTELQQWNELYGEGGSRVKKALSY 938
Score = 68.2 bits (165), Expect = 1e-11
Identities = 32/56 (57%), Positives = 41/56 (73%)
Frame = +2
Query: 340 VVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPP 395
V+PP +IGV FDDIGALE+VK L ELV+LP++RPELF G L + V+G + P
Sbjct: 2 VIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKXVQGYPIIWSP 169
>TC207657 similar to UP|Q6YUV9 (Q6YUV9) Transitional endoplasmic reticulum
ATPase-like, partial (37%)
Length = 919
Score = 328 bits (842), Expect = 3e-90
Identities = 161/182 (88%), Positives = 169/182 (92%)
Frame = +3
Query: 460 ARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVD 519
ARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVD
Sbjct: 9 ARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVD 188
Query: 520 LPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
LPDA+NR KILRI L ENL+ DFQFDKLANLTDGYSGSDLKNLCI AAYRPVQELLEEE
Sbjct: 189 LPDAENRMKILRIFLAQENLNSDFQFDKLANLTDGYSGSDLKNLCIAAAYRPVQELLEEE 368
Query: 580 KAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFG 639
K G +N T LRPLNLDDF+Q+KSKVGPSVAYDA S+NELRKWNEMYGEGG+RTK+PFG
Sbjct: 369 KKGASNDTTSILRPLNLDDFIQAKSKVGPSVAYDATSMNELRKWNEMYGEGGSRTKAPFG 548
Query: 640 FG 641
FG
Sbjct: 549 FG 554
>TC229555 similar to GB|AAM62497.1|21553404|AY085265 26S proteasome
regulatory particle chain RPT6-like protein {Arabidopsis
thaliana;} , partial (85%)
Length = 1155
Score = 283 bits (724), Expect = 2e-76
Identities = 155/337 (45%), Positives = 217/337 (63%), Gaps = 5/337 (1%)
Frame = +2
Query: 300 RESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDV 359
RES + A+ K + +P + + +E ++ P I V+F+ IG LE +
Sbjct: 104 RESSKKALEHKKEIAKRLGRPL-----IQTNPYEDVIACDIINPDHIDVEFNSIGGLETI 268
Query: 360 KKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSG 419
K+AL ELVILP++RP+LF+HG LL P KG+LL+GPPGTGKT++AKA+A E+ A FI+V
Sbjct: 269 KQALFELVILPLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESRAVFINVRI 448
Query: 420 STLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMA 479
S L SKWFGDA+KL A+FS A KL P IIF+DEVDS LG R G +HEA M+ EFMA
Sbjct: 449 SNLMSKWFGDAQKLVAAVFSLAYKLQPAIIFIDEVDSFLGKRRGT-DHEAMLNMKTEFMA 625
Query: 480 AWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENL 539
WDG + +N ++++L ATNRP +LD+A++RRLP+ + +PD R +IL+++LK E +
Sbjct: 626 LWDGFTTDQNAQVMVLAATNRPSELDEAILRRLPQAFEIGIPDQRERAEILKVVLKGERV 805
Query: 540 DPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDF 599
+ + F +A L +GY+GSDL +LC AAY P++ELL+EEK G + A RPL+ DF
Sbjct: 806 EDNIDFGHIAGLCEGYTGSDLFDLCKKAAYFPIRELLDEEKKGKQ---SHAPRPLSQLDF 976
Query: 600 -----VQSKSKVGPSVAYDAVSVNELRKWNEMYGEGG 631
K+KV S Y S+ +W + GE G
Sbjct: 977 EKALATSKKTKVAAS-EYGGFSLQSPSRWT-VPGESG 1081
>TC219888 similar to UP|Q944N4 (Q944N4) Tobacco mosaic virus helicase
domain-binding protein (Fragment), partial (50%)
Length = 1119
Score = 222 bits (566), Expect = 4e-58
Identities = 122/275 (44%), Positives = 185/275 (66%), Gaps = 3/275 (1%)
Frame = +3
Query: 357 EDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFIS 416
E K+AL E+VILP +R +LFT L RP +G+LLFGPPG GKT++AKA+A+E+ A F +
Sbjct: 3 EKAKQALMEMVILPTKRRDLFT--GLRRPARGLLLFGPPGNGKTMLAKAVASESQATFFN 176
Query: 417 VSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNE 476
V+ ++LTSKW G+AEKL + LF A P +IF+DE+DS++ R A E++A+RR+++E
Sbjct: 177 VTAASLTSKWVGEAEKLVRTLFMVAISRQPSVIFIDEIDSIMSTR-LANENDASRRLKSE 353
Query: 477 FMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKH 536
F+ +DG+ S + ++++GATN+P +LDDAV+RRL +RIY+ LPD + RK +L+ LK
Sbjct: 354 FLIQFDGVTSNPDDIVIVIGATNKPQELDDAVLRRLVKRIYIPLPDENVRKLLLKHKLKG 533
Query: 537 ENLD-PDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVA--LRP 593
+ P ++L T+GYSGSDL+ LC AA P++EL G + +T A +R
Sbjct: 534 QAFSLPSRDLERLVKETEGYSGSDLQALCEEAAMMPIREL------GADILTVKANQVRG 695
Query: 594 LNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
L +DF ++ + + PS+ + EL +WNE +G
Sbjct: 696 LRYEDFKKAMATIRPSL--NKSKWEELERWNEEFG 794
>TC208420 homologue to UP|Q7XXR9 (Q7XXR9) Katanin, partial (79%)
Length = 1523
Score = 205 bits (522), Expect = 4e-53
Identities = 115/289 (39%), Positives = 176/289 (60%), Gaps = 4/289 (1%)
Frame = +1
Query: 344 GEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMA 403
G VK++ I LE+ K+ L E V++P++ P+ FT LL P KGILLFGPPGTGKT++A
Sbjct: 442 GSPDVKWESIKGLENAKRLLKEAVVMPIKYPKYFT--GLLSPWKGILLFGPPGTGKTMLA 615
Query: 404 KALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGG 463
KA+ATE F ++S S++ SKW GD+EKL K LF A AP IF+DE+D+++ RG
Sbjct: 616 KAVATECKTTFFNISASSVVSKWRGDSEKLVKVLFELARHHAPSTIFLDEIDAIISQRGE 795
Query: 464 A-FEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPD 522
A EHEA+RR++ E + DGL +K ++ + +L ATN P++LD A++RRL +RI V LP+
Sbjct: 796 ARSEHEASRRLKTELLIQMDGL-TKTDELVFVLAATNLPWELDAAMLRRLEKRILVPLPE 972
Query: 523 ADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAG 582
R+ + +L + + +D L + T+GYSGSD++ LC A +P++ L+ + +
Sbjct: 973 PVARRAMFEELLPQQPDEEPIPYDILVDKTEGYSGSDIRLLCKETAMQPLRRLMSQLEQS 1152
Query: 583 DNNITNVAL---RPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
+ + L P+ +D + PS A ++ K+N YG
Sbjct: 1153QDVVPEEELPKVGPIKSEDIETALRNTRPSAHLHA---HKYDKFNADYG 1290
>TC204689 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (98%)
Length = 1726
Score = 193 bits (491), Expect = 2e-49
Identities = 103/230 (44%), Positives = 148/230 (63%), Gaps = 3/230 (1%)
Frame = +3
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
VK++D+ LE K+AL E VILP++ P+ FT RP + LL+GPPGTGK+ +AKA+A
Sbjct: 564 VKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 737
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
TEA + F SVS S L SKW G++EKL LF A + AP IIF+DE+DSL G RG E
Sbjct: 738 TEAESTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFIDEIDSLCGQRGEGNES 917
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRK 527
EA+RR++ E + G+ +Q++L+L ATN P+ LD A+ RR +RIY+ LPD R+
Sbjct: 918 EASRRIKTELLVQMQGV-GHNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQ 1094
Query: 528 KILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE 574
+ ++ L H + DF++ LA+ T+G+SGSD+ + PV++
Sbjct: 1095HMFKVHLGDTPHNLTESDFEY--LASRTEGFSGSDISVCVKDVLFEPVRK 1238
>TC204690 homologue to UP|Q9SEA8 (Q9SEA8) Salt-induced AAA-Type ATPase,
partial (94%)
Length = 1630
Score = 192 bits (489), Expect = 3e-49
Identities = 116/314 (36%), Positives = 178/314 (55%), Gaps = 30/314 (9%)
Frame = +2
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
VK++D+ LE K+AL E VILP++ P+ FT RP + LL+GPPGTGK+ +AKA+A
Sbjct: 515 VKWNDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 688
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
TEA + F SVS S L SKW G++EKL LF A + AP IIFVDE+DSL G RG E
Sbjct: 689 TEADSTFFSVSSSDLVSKWMGESEKLVSNLFQMARESAPSIIFVDEIDSLCGQRGEGNES 868
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRK 527
EA+RR++ E + G+ +Q++L+L ATN P+ LD A+ RR +RIY+ LPD R+
Sbjct: 869 EASRRIKTELLVQMQGV-GHNDQKVLVLAATNTPYALDQAIRRRFDKRIYIPLPDLKARQ 1045
Query: 528 KILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLE------- 577
+ ++ L H + D F+ LA T+G+SGSD+ + PV++ +
Sbjct: 1046HMFKVHLGDTPHNLAESD--FEHLARKTEGFSGSDISVCVKDVLFEPVRKTQDAMFFFRN 1219
Query: 578 -----------EEKAGDNNITNVALR---------PLNLDDFVQSKSKVGPSVAYDAVSV 617
++ A + ++A + P++ DF + ++ P+V+ + V
Sbjct: 1220PEDMWIPCGPKQQSAVQTTMQDLAAKGLASKILPPPISRTDFDKVLARQRPTVSKSDLDV 1399
Query: 618 NELRKWNEMYGEGG 631
+E ++ + +GE G
Sbjct: 1400HE--RFTKEFGEEG 1435
>TC221009 homologue to UP|Q940D1 (Q940D1) At1g64110/F22C12_22, partial (18%)
Length = 696
Score = 184 bits (468), Expect = 8e-47
Identities = 85/143 (59%), Positives = 115/143 (79%)
Frame = +1
Query: 438 FSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGA 497
F+ A+K++P IIFVDEVDS+LG R EHEA R+++NEFM WDGL +K+ +RIL+L A
Sbjct: 1 FTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLLTKQGERILVLAA 180
Query: 498 TNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSG 557
TNRPFDLD+A+IRR RRI V LP +NR+KILR +L E +D + +F ++A +T+GY+G
Sbjct: 181 TNRPFDLDEAIIRRFERRIMVGLPSVENREKILRTLLAKEKVDNELEFKEIATMTEGYTG 360
Query: 558 SDLKNLCITAAYRPVQELLEEEK 580
SDLKNLC TAAYRPV+EL+++E+
Sbjct: 361 SDLKNLCTTAAYRPVRELIQQER 429
>TC206116 UP|CC48_SOYBN (P54774) Cell division cycle protein 48 homolog
(Valosin containing protein homolog) (VCP), complete
Length = 2583
Score = 170 bits (431), Expect = 2e-42
Identities = 93/236 (39%), Positives = 142/236 (59%), Gaps = 4/236 (1%)
Frame = +1
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LE+VK+ L E V P+ PE F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 1453 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGM-SPSKGVLFYGPPGCGKTLLAKAIA 1629
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFISV G L + WFG++E + +F A + AP ++F DE+DS+ RG +
Sbjct: 1630 NECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGSSVGD 1809
Query: 468 E--ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+ + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 1810 AGGAADRVLNQLLTEMDGMSAKKT--VFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDE 1983
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
D+R +I + L+ + + LA T G+SG+D+ +C A ++E +E++
Sbjct: 1984 DSRHQIFKACLRKSPIAKNVDLRALARHTQGFSGADITEICQRACKYAIRENIEKD 2151
Score = 165 bits (417), Expect = 7e-41
Identities = 101/285 (35%), Positives = 157/285 (54%), Gaps = 4/285 (1%)
Frame = +1
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DD+G + + ELV LP+R P+LF + +P KGILL+GPPG+GKTL+A+A+A
Sbjct: 634 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGV-KPPKGILLYGPPGSGKTLIARAVA 810
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+DS+ R
Sbjct: 811 NETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HG 987
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL+S+ + ++++GATNRP +D A+ R R R I + +PD
Sbjct: 988 EVERRIVSQLLTLMDGLKSRAH--VIVIGATNRPNSIDPALRRFGRFDREIDIGVPDEVG 1161
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R ++LRI K+ L D +++A T GY G+DL LC AA + ++E ++ D
Sbjct: 1162RLEVLRIHTKNMKLSDDVDLERIAKDTHGYVGADLAALCTEAALQCIREKMDVIDLEDET 1341
Query: 586 ITNVALRPLNL--DDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I L + + + F + PS + V W ++ G
Sbjct: 1342IDAEVLNSMAVTNEHFQTALGTSNPSALRETVVEVPNVSWEDIGG 1476
>BU578832
Length = 437
Score = 170 bits (431), Expect = 2e-42
Identities = 86/149 (57%), Positives = 116/149 (77%), Gaps = 3/149 (2%)
Frame = +1
Query: 409 EAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHE 468
EAGA+FI+VS S +TSKWFG+ EK +ALFS A+K+AP IIF+DEVDS+LG R EHE
Sbjct: 1 EAGASFINVSISNITSKWFGEDEKNVRALFSLAAKVAPTIIFIDEVDSMLGKRTKYGEHE 180
Query: 469 ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKK 528
A R+++NEFMA WDG+ +K +RIL+L ATNRPFDLD+A+IRR RRI V LP A+NR+
Sbjct: 181 AMRKIKNEFMAHWDGILTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREM 360
Query: 529 ILRIIL---KHENLDPDFQFDKLANLTDG 554
IL+ +L K+E++D F++L+ +T+G
Sbjct: 361 ILKTLLAKEKYEHID----FNELSTITEG 435
>BE191235
Length = 313
Score = 163 bits (412), Expect = 3e-40
Identities = 82/102 (80%), Positives = 93/102 (90%), Gaps = 2/102 (1%)
Frame = +3
Query: 169 MILPNFARLAKLPLSLKRLTD--KGQKTSDDDEINKLFSNVVSIQPPKDENLLATFKKQL 226
MILPNF R+AKLPLSLKRLT+ KG KTS+DDEINKLFSNV+S+ PPKDENLLATFKKQL
Sbjct: 3 MILPNFGRVAKLPLSLKRLTEGIKGDKTSEDDEINKLFSNVLSMHPPKDENLLATFKKQL 182
Query: 227 EEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTK 268
EED+KIV SRSNLN LRKVLEEHQLSC+DLLH+++D + LTK
Sbjct: 183 EEDKKIVTSRSNLNVLRKVLEEHQLSCMDLLHVNTDGIFLTK 308
>BI893296 similar to GP|21553404|gb 26S proteasome regulatory particle chain
RPT6-like protein {Arabidopsis thaliana}, partial (32%)
Length = 398
Score = 160 bits (405), Expect = 2e-39
Identities = 80/133 (60%), Positives = 99/133 (74%)
Frame = +1
Query: 356 LEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFI 415
LE +K AL ELVILP++RP+LF+HG LL P KG+LL+GPPGTGKT++AKA+A E+GA FI
Sbjct: 1 LETIKLALFELVILPLKRPDLFSHGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFI 180
Query: 416 SVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRN 475
+V L SKWFGDA+KL A+F A KL P IIF+DEVDS LG R +HEA M+
Sbjct: 181 NVRIFFLMSKWFGDAQKLVTAIFXLAHKLQPAIIFIDEVDSFLGQR-RTTDHEALLNMKT 357
Query: 476 EFMAAWDGLRSKE 488
EFMA WDG + +
Sbjct: 358 EFMALWDGFTTDQ 396
>TC214213 homologue to UP|PRSA_BRACM (O23894) 26S protease regulatory subunit
6A homolog (TAT-binding protein homolog 1) (TBP-1),
partial (64%)
Length = 1086
Score = 147 bits (370), Expect = 2e-35
Identities = 88/225 (39%), Positives = 131/225 (58%), Gaps = 7/225 (3%)
Frame = +1
Query: 350 FDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATE 409
++DIG LE + L E ++LPM E F + RP KG+LL+GPPGTGKTLMA+A A +
Sbjct: 46 YNDIGGLEKQIQELVEAIVLPMTHKERFQKLGV-RPPKGVLLYGPPGTGKTLMARACAAQ 222
Query: 410 AGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEA 469
A F+ ++G L + GD KL + F A + +P IIF+DE+D++ R F+ E
Sbjct: 223 TNATFLKLAGPQLVQMFIGDGAKLVRDAFQLAKEKSPCIIFIDEIDAIGTKR---FDSEV 393
Query: 470 T-----RRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPD 522
+ +R E + DG S + RI ++ ATNR LD A++R RL R+I P
Sbjct: 394 SGDREVQRTMLELLNQLDGFSS--DDRIKVIAATNRADILDPALMRSGRLDRKIEFPHPS 567
Query: 523 ADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITA 567
+ R +IL+I + N+ PD F++LA TD ++G+ LK +C+ A
Sbjct: 568 EEARARILQIHSRKMNVHPDVNFEELARSTDDFNGAQLKAVCVEA 702
>TC216547
Length = 733
Score = 145 bits (367), Expect = 4e-35
Identities = 90/219 (41%), Positives = 133/219 (60%), Gaps = 5/219 (2%)
Frame = +2
Query: 181 PLSLKRLTDKGQKTSDDDE-INKLFSNVVSIQPPKDENLLATFKKQLEEDRKIVISRSNL 239
P S RL D+G++ ++ + KLF N V+I P+DE LLA++K+QL+ D + + + NL
Sbjct: 77 PDSFGRLHDRGKEAPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKENL 256
Query: 240 NELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGD---KL 296
+ LR VL + C L L + L+ + AEK+VGWA LS L+ + D KL
Sbjct: 257 HNLRTVLSRCGVECEGLETLCIRNQTLSIENAEKIVGWA----LSCHLMQNAETDPDAKL 424
Query: 297 CLPRESLEIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGA 355
L +S++ + L + S+ +SLK++ ++EFE ++ V+PP +IGV FDDIGA
Sbjct: 425 VLSCKSIQYGVGILHATQNESKSLKKSLKDVVTENEFEKRLLADVIPPNDIGVTFDDIGA 604
Query: 356 LEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGP 394
LE+VK L ELV+LP++RPELF G L +P GILL P
Sbjct: 605 LENVKDTLKELVMLPLQRPELFCKGQLXQPCXGILLLVP 721
>TC204912 weakly similar to GB|AAH46286.1|28279482|BC046286 spastic
paraplegia 4 homolog {Mus musculus;} , partial (4%)
Length = 911
Score = 145 bits (366), Expect = 5e-35
Identities = 72/141 (51%), Positives = 104/141 (73%), Gaps = 11/141 (7%)
Frame = +2
Query: 504 LDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNL 563
LD+AVIRRLPRR+ V+LPDA NR+KI+ +IL E L PD F+ +AN+TDGYSGSDLKNL
Sbjct: 14 LDEAVIRRLPRRLMVNLPDAPNREKIVSVILAKEELAPDVDFEAIANMTDGYSGSDLKNL 193
Query: 564 CITAAYRPVQELLEEEK-----AGDNN------ITNVALRPLNLDDFVQSKSKVGPSVAY 612
C+TAA+ P++E+LE+EK A N ++ +RPL ++DF+ + +V SV+
Sbjct: 194 CVTAAHCPIREILEKEKKERSLALTENQPLPQLCSSTDIRPLKMEDFIYAHEQVCVSVSS 373
Query: 613 DAVSVNELRKWNEMYGEGGTR 633
++ ++NEL +WN++YGEGG+R
Sbjct: 374 ESTNMNELLQWNDLYGEGGSR 436
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,407,760
Number of Sequences: 63676
Number of extensions: 311755
Number of successful extensions: 1731
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 1655
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1669
length of query: 645
length of database: 12,639,632
effective HSP length: 103
effective length of query: 542
effective length of database: 6,081,004
effective search space: 3295904168
effective search space used: 3295904168
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0135.9


