

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
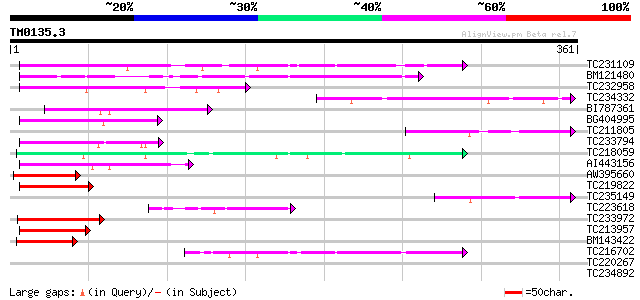
Query= TM0135.3
(361 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 105 3e-23
BM121480 67 1e-11
TC232958 67 2e-11
TC234332 64 8e-11
BI787361 64 8e-11
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 63 2e-10
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 63 2e-10
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 62 4e-10
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 62 5e-10
AI443156 59 4e-09
AW395660 58 6e-09
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 57 1e-08
TC235149 52 4e-07
TC223618 50 2e-06
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 48 8e-06
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 48 8e-06
BM143422 47 2e-05
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 46 2e-05
TC220267 similar to UP|Q6K235 (Q6K235) F-box-like protein, parti... 40 0.001
TC234892 weakly similar to UP|Q7QIA5 (Q7QIA5) AgCP3966 (Fragment... 36 0.024
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 105 bits (262), Expect = 3e-23
Identities = 84/299 (28%), Positives = 145/299 (48%), Gaps = 14/299 (4%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 66
LP E+ EILS LPVKS++R R K W+SII F+ HL++S S+ ++L SL
Sbjct: 93 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKSHSSLILRHRSHLYSL 272
Query: 67 -ITSPRKS---RSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRS- 121
+ SP ++ S + + G+ NGL+C+ + D + LWNP +R
Sbjct: 273 DLKSPEQNPVELSHPLMCYSNSIKVLGSSNGLLCISNVADD--------IALWNPFLRKH 428
Query: 122 -------MSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVA--VALSWDTLETMVNVYN 172
+ L+ VY GF + S ++ YK++++ V L T ++ V +Y
Sbjct: 429 RILPADRFHRPQSSLFAARVY----GFGHHSPSNDYKLLSITYFVDLQKRTFDSQVQLYT 596
Query: 173 IGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSFDFGKESCA 232
+ D+ W+ +P S ++ V+V+ +L+ L + ++ + I+SFD +E+
Sbjct: 597 LKSDS-WKNLP-SMPYALCCARTMGVFVSGSLHWL-VTRKLQPHEPDLIVSFDLTRETFH 767
Query: 233 QLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWTLLFNI 291
++PLP +D + +L CLC+ + + F VW M+ +G SW LF +
Sbjct: 768 EVPLPVTVNGDFDM-----QVALLGGCLCVVE-HRGTGFDVWVMRVYGSRNSWEKLFTL 926
>BM121480
Length = 917
Score = 67.0 bits (162), Expect = 1e-11
Identities = 70/259 (27%), Positives = 114/259 (43%), Gaps = 2/259 (0%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 66
LP+EL EIL LPVKSL+RF+CV +K+ D + L L S R DF +
Sbjct: 66 LPQELIREILLRLPVKSLLRFKCV--LFKTCSRDVVYFPLPL-PSIPCLRLDDFGIRPKI 236
Query: 67 ITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRS 126
+ G+C GLV L+ YD ++++ LWNP++ +
Sbjct: 237 L---------------------GSCRGLVLLY---YDD----SANLILWNPSLG--RHKX 326
Query: 127 PPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDTCWRTIPVSH 186
P Y + +GF YD S D Y + + + L ET ++Y+ ++ W+T + +
Sbjct: 327 LPNYRDDITSFPYGFGYDESKDEY--LLILIGLPKFGPETGADIYSFKTES-WKTDTIVY 497
Query: 187 LP-SMYLKDDDAVYVNNTLN-RLAILPNVEDRDRCTILSFDFGKESCAQLPLPYCPQSRY 244
P Y +D + LN L E + I++FD + + + +PLP +S
Sbjct: 498 DPLXRYXAEDXIARAGSLLNGALHWFVFSESKXDHVIIAFDLVERTLSXIPLPLADRSTV 677
Query: 245 DFYRTWPTLGVLRDCLCIC 263
+ L ++ CL +C
Sbjct: 678 QKDAVY-GLKIMGXCLXVC 731
>TC232958
Length = 758
Score = 66.6 bits (161), Expect = 2e-11
Identities = 53/166 (31%), Positives = 72/166 (42%), Gaps = 19/166 (11%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLH-----------LHRSSSTT 55
LP++L EIL LPVKSLVRF+ V KSW +ISD +F K H L +SS
Sbjct: 61 LPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIASSAP 240
Query: 56 RNTDFAYLQSLITSPRKSRSASTIAIPDDY----DFCGTCNGLVCLHSSKYDRKDKYTSH 111
+ SL + P Y + G+C G + LH SH
Sbjct: 241 ELRSIDFNASLHDDSASVAVTVDLPAPKPYFHFVEIIGSCRGFILLHC---------LSH 393
Query: 112 VRLWNP--AMRSMSQRSPPLYL--PIVYDSYFGFDYDSSTDTYKVV 153
+ +WNP + + SP + + + GF YD STD + VV
Sbjct: 394 LCVWNPTTGVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDDFLVV 531
>TC234332
Length = 644
Score = 64.3 bits (155), Expect = 8e-11
Identities = 55/179 (30%), Positives = 78/179 (42%), Gaps = 14/179 (7%)
Frame = +2
Query: 196 DAVYVNNTLNRLAILPNVEDR-------DRCTILSFDFGKESCAQLPLPYCPQSRYDFYR 248
D V T+N LA+ + D D I S+D ES L +P +
Sbjct: 17 DGASVRGTVNWLALPNSSSDYQWETVTIDDLVIFSYDLKNESYRYLLMP---DGLLEVPH 187
Query: 249 TWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWTLLFNIAGYAYDIPCRFF----A 304
+ P L VL+ CLC+ + HF W MKEFG+ KSWT NI+ I F
Sbjct: 188 SPPELVVLKGCLCLSHRHGGNHFGFWLMKEFGVEKSWTRFLNISYDQLHIHGGFLDHPVI 367
Query: 305 MYMSENGDALLLSKLAVSQPQAVLYTQKDNKLEV---TDVANYIFNCYAMNYIESLVPP 360
+ MSE+ +LL + +LY ++DN +E D + F Y +Y +S V P
Sbjct: 368 LCMSEDDGVVLLEN--GGHGKFILYNKRDNTIECYGELDKGRFQFLSY--DYAQSFVMP 532
>BI787361
Length = 423
Score = 64.3 bits (155), Expect = 8e-11
Identities = 41/124 (33%), Positives = 58/124 (46%), Gaps = 17/124 (13%)
Frame = +3
Query: 23 SLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTR---------NTDFAY--------LQS 65
+L+RFR VS++W S+I D F+KLHL RS T + D ++
Sbjct: 3 ALMRFRYVSETWNSLIFDPTFVKLHLERSPKNTHVLLEFQAIYDRDVGQQVGVAPCSIRR 182
Query: 66 LITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQR 125
L+ +P + G+CNGLVC+ R+ + RLWNPA MS+
Sbjct: 183 LVENPSFTIDDCLTLFKHTNSIFGSCNGLVCMTKCFDVREFEEECQYRLWNPATGIMSEY 362
Query: 126 SPPL 129
SPPL
Sbjct: 363 SPPL 374
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 63.2 bits (152), Expect = 2e-10
Identities = 38/99 (38%), Positives = 49/99 (49%), Gaps = 8/99 (8%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNT-------- 58
LP E+ EILS LPV+SL+RFR SKSWKS+I LHL RS + NT
Sbjct: 40 LPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLASNTSLILRVDS 219
Query: 59 DFAYLQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCL 97
D P S + + + G+CNGL+C+
Sbjct: 220 DLYQTNFPTLDPPVSLNHPLMCYSNSITLLGSCNGLLCI 336
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 62.8 bits (151), Expect = 2e-10
Identities = 39/114 (34%), Positives = 58/114 (50%), Gaps = 6/114 (5%)
Frame = +1
Query: 253 LGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWTLLFNI------AGYAYDIPCRFFAMY 306
LGVL+ CLC+ K HFVVW MK++G +SW L +I ++Y P Y
Sbjct: 4 LGVLQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVPNPENFSYSGP-----YY 168
Query: 307 MSENGDALLLSKLAVSQPQAVLYTQKDNKLEVTDVANYIFNCYAMNYIESLVPP 360
+SENG+ LL+ + +LY +DN + + + A Y+E+LV P
Sbjct: 169 ISENGEVLLMFEF-----DLILYNPRDNSFKYPKIESGKGWFDAEVYVETLVSP 315
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 62.0 bits (149), Expect = 4e-10
Identities = 47/113 (41%), Positives = 62/113 (54%), Gaps = 21/113 (18%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTT-----RNTDFA 61
LP+EL EIL LPVKSL+RF+CV KS+ S+ISD QF+ H ++S T R+ DF
Sbjct: 109 LPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAASPTHRLILRSHDF- 285
Query: 62 YLQSLIT-SPRKSRSASTIAIP-----------DDY----DFCGTCNGLVCLH 98
Y QS+ T S K+ S + P DD+ G+C GLV L+
Sbjct: 286 YAQSIATESVFKTCSRDVVYFPLPLPSIPCLRLDDFGIRPKILGSCRGLVLLY 444
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 61.6 bits (148), Expect = 5e-10
Identities = 73/321 (22%), Positives = 124/321 (37%), Gaps = 34/321 (10%)
Frame = +3
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIK----------------LHL 48
+ LP EL +LS LP K L+ +CV KSW +I+D F+ L +
Sbjct: 60 EHLPGELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHLLVI 239
Query: 49 HRSSSTTRNTDFAYLQSLITSPRKSRSASTIAIPDDY--------DFCGTCNGLVCLHSS 100
R + T + L P+K S+ + P +Y + G CNG+ L +
Sbjct: 240 RRPFFSGLKTYISVLSWNTNDPKKHVSSDVLNPPYEYNSDHKYWTEILGPCNGIYFLEGN 419
Query: 101 KYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALS 160
+ + A+ SP + + Y GF +D T+ YKVV +
Sbjct: 420 PNVLMNPSLGEFK----ALPKSHFTSP--HGTYTFTDYAGFGFDPKTNDYKVVVLKDLWL 581
Query: 161 WDTLETMV-----NVYNIGDDTCWRTIPVSHLP---SMYLKDDDAVYVNNTLNRLAILPN 212
+T E + +Y++ ++ WR + S LP ++ Y NN + +
Sbjct: 582 KETDEREIGYWSAELYSLNSNS-WRKLDPSLLPLPIEIWGSSRVFTYANNCCHWWGFVEE 758
Query: 213 VEDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTL--GVLRDCLCICQKDKNMH 270
D + +L+FD KES ++ +P S + + T L +
Sbjct: 759 -SDATQDVVLAFDMVKESFRKIRVPKIRDSSDEKFGTLVPFEESASIGFLVYPVRGTEKR 935
Query: 271 FVVWQMKEFGLYKSWTLLFNI 291
F VW MK++ SW +++
Sbjct: 936 FDVWVMKDYWDEGSWVKQYSV 998
>AI443156
Length = 414
Score = 58.5 bits (140), Expect = 4e-09
Identities = 41/116 (35%), Positives = 57/116 (48%), Gaps = 5/116 (4%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRS--SSTTRNTDFAY-- 62
LP E+ EILS LPVKS++R R K W+SII FI HL++S S R+ Y
Sbjct: 90 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFHLNKSHTSLILRHRSQLYSL 269
Query: 63 -LQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNP 117
L+SL+ S + + G+ NGL+C+ + D + LWNP
Sbjct: 270 DLKSLLDPNPFELSHPLMCYSNSIKVLGSSNGLLCISNVXDD--------IALWNP 413
>AW395660
Length = 381
Score = 58.2 bits (139), Expect = 6e-09
Identities = 28/43 (65%), Positives = 34/43 (78%)
Frame = +3
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIK 45
P FLP+EL +EILS LPVKSL++FRCV KSW S+I D F+K
Sbjct: 252 PLPFLPDELVVEILSRLPVKSLLQFRCVCKSWMSLIYDPYFMK 380
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 57.0 bits (136), Expect = 1e-08
Identities = 25/47 (53%), Positives = 33/47 (70%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS 53
LPE L +EIL W+ V + ++ RCV K WKS++ D QF+K HLH S S
Sbjct: 196 LPEGLMIEILVWIRVSNPLQLRCVCKRWKSLVVDPQFVKKHLHTSLS 336
>TC235149
Length = 435
Score = 52.0 bits (123), Expect = 4e-07
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 7/97 (7%)
Frame = +2
Query: 271 FVVWQMKEFGLYKSWTLLFNIA-----GYAYDIPCRFFA-MYMSENGDALLLSKLAVSQP 324
FVVW +EFG+ +SWT L N++ + RF + MSEN D LLL+ S
Sbjct: 2 FVVWLTREFGVERSWTRLLNVSYEHFRNHGCPPYYRFVTPLCMSENEDVLLLANDEGS-- 175
Query: 325 QAVLYTQKDNKLE-VTDVANYIFNCYAMNYIESLVPP 360
+ V Y +DN+++ + D +Y F+ + +Y+ SLV P
Sbjct: 176 EFVFYNLRDNRIDRIQDFDSYKFSFLSHDYVPSLVLP 286
>TC223618
Length = 444
Score = 50.1 bits (118), Expect = 2e-06
Identities = 38/97 (39%), Positives = 55/97 (56%), Gaps = 3/97 (3%)
Frame = +1
Query: 89 GTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSPPL---YLPIVYDSYFGFDYDS 145
G+CNGL+C + K D V LWNP++R +S++SPPL + P + + FG YD
Sbjct: 133 GSCNGLLCF-AIKGDC-------VLLWNPSIR-VSKKSPPLGNNWRPGCFTA-FGLGYDH 282
Query: 146 STDTYKVVAVAVALSWDTLETMVNVYNIGDDTCWRTI 182
+ YKVVAV S +E V VY++ ++ WR I
Sbjct: 283 VNEDYKVVAVFCDPSEYFIECKVKVYSMATNS-WRKI 390
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 47.8 bits (112), Expect = 8e-06
Identities = 26/55 (47%), Positives = 35/55 (63%)
Frame = -2
Query: 6 FLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDF 60
FLP+E+ +EIL LPVKSL F+ V KS S+IS+ F K H R+++ T F
Sbjct: 543 FLPQEIIIEILLRLPVKSLPSFKFVCKS*LSLISNPHFAKWHFERNAAHTEKLLF 379
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 47.8 bits (112), Expect = 8e-06
Identities = 25/45 (55%), Positives = 30/45 (66%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRS 51
LP EL EIL PV+S++RF+CV KSW S+ISD QF L S
Sbjct: 33 LPLELIREILLRSPVRSVLRFKCVCKSWLSLISDPQFTHFDLAAS 167
>BM143422
Length = 424
Score = 46.6 bits (109), Expect = 2e-05
Identities = 24/39 (61%), Positives = 28/39 (71%)
Frame = +1
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQF 43
Q LP +L IL LPVKS+ RF+CV KSW S+ISD QF
Sbjct: 223 QTLPLDLIELILLKLPVKSVTRFKCVCKSWLSLISDPQF 339
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 46.2 bits (108), Expect = 2e-05
Identities = 49/193 (25%), Positives = 83/193 (42%), Gaps = 13/193 (6%)
Frame = +3
Query: 112 VRLWNPAMRSMSQRSPPLYLPIVYDSY----------FGFDYDSSTDTYKVVAVA--VAL 159
+ WNP++R R P YLP+ + GF +D T YK+V ++ V L
Sbjct: 30 IAFWNPSLRQ--HRILP-YLPVPRRRHPDTTLFAARVCGFGFDHKTRDYKLVRISYFVDL 200
Query: 160 SWDTLETMVNVYNIGDDTCWRTIPVSHLP-SMYLKDDDAVYVNNTLNRLAILPNVEDRDR 218
+ + V +Y + + W+T+P LP ++ V+V N+L+ + + +E
Sbjct: 201 HDRSFDAQVKLYTLRANA-WKTLP--SLPYALCCARTMGVFVGNSLHWV-VTRKLEPDQP 368
Query: 219 CTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKE 278
I++FD + +LPLP F L +L LC+ VW M+E
Sbjct: 369 DLIIAFDLTHDIFRELPLPDTGGVDGGFEID---LALLGGSLCMTVNFHKTRIDVWVMRE 539
Query: 279 FGLYKSWTLLFNI 291
+ SW +F +
Sbjct: 540 YNRRDSWCKVFTL 578
>TC220267 similar to UP|Q6K235 (Q6K235) F-box-like protein, partial (7%)
Length = 586
Score = 40.4 bits (93), Expect = 0.001
Identities = 16/38 (42%), Positives = 26/38 (68%)
Frame = +2
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFI 44
LP++L IL++LP+ S+ R CVSK W I++ +F+
Sbjct: 338 LPDDLLERILAYLPIASIFRAGCVSKRWHEIVNSERFV 451
>TC234892 weakly similar to UP|Q7QIA5 (Q7QIA5) AgCP3966 (Fragment), partial
(4%)
Length = 370
Score = 36.2 bits (82), Expect = 0.024
Identities = 18/49 (36%), Positives = 31/49 (62%)
Frame = -2
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTT 55
LP+EL I L + ++R + ++KS+ IISD+ F++ + S+STT
Sbjct: 243 LPQELIQNIFLSLVLPEIIRLKLLNKSFSRIISDNTFVRQYNSLSTSTT 97
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,467,034
Number of Sequences: 63676
Number of extensions: 332928
Number of successful extensions: 1672
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1658
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1664
length of query: 361
length of database: 12,639,632
effective HSP length: 98
effective length of query: 263
effective length of database: 6,399,384
effective search space: 1683037992
effective search space used: 1683037992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0135.3


