

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.13
(479 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
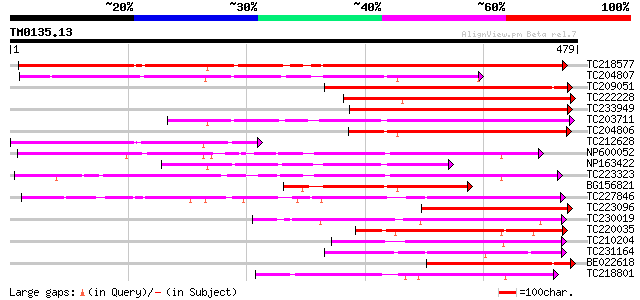
Score E
Sequences producing significant alignments: (bits) Value
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 436 e-122
TC204807 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone g... 290 1e-78
TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 247 7e-66
TC222228 weakly similar to UP|HQGT_ARATH (Q9M156) Probable hydro... 240 1e-63
TC233949 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 219 3e-57
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 213 1e-55
TC204806 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone g... 196 3e-50
TC212628 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 168 4e-42
NP600052 zeatin O-glucosyltransferase [Glycine max] 164 8e-41
NP163422 UDP-glycose:flavonoid glycosyltransferase 163 1e-40
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 161 7e-40
BG156821 similar to SP|Q9AR73|HQGT Hydroquinone glucosyltransfer... 160 9e-40
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 159 2e-39
TC223096 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 157 8e-39
TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferas... 155 4e-38
TC220035 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-g... 148 5e-36
TC210204 similar to UP|Q9SY84 (Q9SY84) F14N23.30, partial (21%) 145 3e-35
TC231164 weakly similar to UP|UFO2_MANES (Q40285) Flavonol 3-O-g... 145 4e-35
BE022618 weakly similar to GP|19911209|db glucosyltransferase-13... 144 9e-35
TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, ... 144 1e-34
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 436 bits (1121), Expect = e-122
Identities = 233/471 (49%), Positives = 313/471 (65%), Gaps = 7/471 (1%)
Frame = +1
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSM 67
H+A+V P ++H ILEF KRL HLH + HITC P+V SS KA + LPP I S+
Sbjct: 4 HVAVVPSPGFTHLVPILEFSKRLLHLHPEFHITCFIPSVGSSPTSSKAYVQTLPPTITSI 183
Query: 68 FLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVL 127
FLPP+ LD + + + + + ++ SLP I LK+L CS + ++VVDV L
Sbjct: 184 FLPPITLDHVSDPSVLALQIELSVNLSLPYIREELKSL-CSRAKV--VALVVDVFANGAL 354
Query: 128 PMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCET------VEIPGCMLIHTTDLPD 181
A ELN+LSY+Y +AMLLSL Y + D+ +S E+ ++IPGC+ IH DLP
Sbjct: 355 NFAKELNLLSYIYLPQSAMLLSLYFYSTKLDEILSSESRELQKPIDIPGCVPIHNKDLPL 534
Query: 182 PVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQEN-KGTSSTSIPHVY 240
P + YK FLE +KRF++ DGV +N+F +LE RAL+E+ KG P +Y
Sbjct: 535 PFHDLSGLG--YKGFLERSKRFHVPDGVFMNTFLELESGAIRALEEHVKGK-----PKLY 693
Query: 241 PVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALG 300
PVGP++Q ES G+E E + WLDKQE NSVLYVSFGS GTL+ +QFNELA G
Sbjct: 694 PVGPIIQM------ESIGHENGV-ECLTWLDKQEPNSVLYVSFGSGGTLSQEQFNELAFG 852
Query: 301 LELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEIL 360
LELSG KFLWV+R P + G L +DPLEFLP+GFLERTK QG VVP WA QI++L
Sbjct: 853 LELSGKKFLWVVRAPSGVVSAGYLCAETKDPLEFLPHGFLERTKKQGLVVPSWAPQIQVL 1032
Query: 361 GHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGI 420
GH A GGFL HCGWNS+LES + G+P++ WPL+A+Q + AA+ +D LKVALRPK NE G+
Sbjct: 1033GHSATGGFLSHCGWNSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKVALRPKVNESGL 1212
Query: 421 VGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLA 471
V R +A+V++ ++ +E EI++RM L+ A +A+KEDGSST+T++++A
Sbjct: 1213VEREEIAKVVRGLMGDKESLEIRKRMGLLKIAAANAIKEDGSSTKTLSEMA 1365
>TC204807 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (79%)
Length = 1673
Score = 290 bits (741), Expect = 1e-78
Identities = 170/405 (41%), Positives = 239/405 (58%), Gaps = 13/405 (3%)
Frame = +2
Query: 9 IAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSMF 68
+A++ P H ++EF KR+ H ++ ++ + PT KA+ E LP +I F
Sbjct: 269 VAMLPSPGMGHLIPMIEFAKRVVCYH-NLAVSFVIPTDGPPSKAQKAVLEALPDSISHTF 445
Query: 69 LPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVLP 128
LPPVNL D P I L+ T+ SLPS+ A +L ++ L+++VVD+ T
Sbjct: 446 LPPVNLSDFPPDTKIETLISHTVLRSLPSLRQAFHSLSATNT---LSAVVVDLFSTDAFD 616
Query: 129 MANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCE------TVEIPGCMLIHTTDLPDP 182
+A E N YV++ STA +LSL + T D+ + CE V IPGC+ + DL DP
Sbjct: 617 VAAEFNASPYVFYPSTATVLSLFFHLPTLDQQVQCEFRDLPEPVSIPGCIPLPGKDLLDP 796
Query: 183 VQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPV 242
VQ+R +E YK L KR+ A+G+I NSF +LEP + LQ+ + P VY V
Sbjct: 797 VQDRK--NEAYKWILHHCKRYKEAEGIIGNSFEELEPGAWNELQKEE----QGRPPVYAV 958
Query: 243 GPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLE 302
GPLV+ + + E +RWLD+Q + SVL+VSFGS GTL+ Q NELALGLE
Sbjct: 959 GPLVRMEAGQADS---------ECLRWLDEQPRGSVLFVSFGSGGTLSSAQINELALGLE 1111
Query: 303 LSGVKFLWVIRPPQKLEMIGDLSV----GHEDPLEFLPNGFLERTKGQGFVVPYWADQIE 358
S +FLWV++ P E I + + DPL+FLP GF+ERTKG+GF+V WA Q +
Sbjct: 1112KSEQRFLWVVKSPN--EEIANATYFSAESQADPLQFLPEGFVERTKGRGFLVQSWAPQPQ 1285
Query: 359 ILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYA---DQKMIA 400
+LGH + GGFL HCGWNS+LES ++G+P +AWPL+A DQ++ A
Sbjct: 1286VLGHPSTGGFLTHCGWNSILESVVNGVPFIAWPLFAEPEDQRLYA 1420
>TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (35%)
Length = 1017
Score = 247 bits (631), Expect = 7e-66
Identities = 120/209 (57%), Positives = 156/209 (74%)
Frame = +2
Query: 267 MRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSV 326
+ WLDKQ SVLYVSFGS GTL+ +Q NELA GLELSG +FLWV+R P L
Sbjct: 14 LSWLDKQPPCSVLYVSFGSGGTLSQNQINELASGLELSGQRFLWVLRAPSNSVSAAYLEA 193
Query: 327 GHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIP 386
EDPL+FLP+GFLERTK +G VV WA Q+++LGH ++GGFL HCGWNS LES G+P
Sbjct: 194 SKEDPLQFLPSGFLERTKEKGLVVASWAPQVQVLGHNSVGGFLSHCGWNSTLESVQEGVP 373
Query: 387 IVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRM 446
++ WPL+A+Q+M A + +DGLKVALRPK NE GIV + +A+VIK ++ GEEG +++RM
Sbjct: 374 LITWPLFAEQRMNAVMLTDGLKVALRPKFNEDGIVEKEEIAKVIKCLMDGEEGIGMRERM 553
Query: 447 KRLQDVAIDALKEDGSSTRTITQLALKWK 475
L+D A ALK DGSS++T++QLA +W+
Sbjct: 554 GNLKDSAASALK-DGSSSQTLSQLASQWE 637
>TC222228 weakly similar to UP|HQGT_ARATH (Q9M156) Probable hydroquinone
glucosyltransferase (Arbutin synthase) , partial (38%)
Length = 678
Score = 240 bits (612), Expect = 1e-63
Identities = 112/199 (56%), Positives = 151/199 (75%), Gaps = 3/199 (1%)
Frame = +1
Query: 283 FGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHED---PLEFLPNGF 339
FGS GT + +Q +ELALGLELSG +FLWV+RPP + L ++D PL+FLP+GF
Sbjct: 4 FGSGGTXSQEQMDELALGLELSGHRFLWVLRPPSSVANAAYLGGANDDGVDPLKFLPSGF 183
Query: 340 LERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMI 399
LERTKGQG VVP WA Q+++LGH ++GGFL HCGWNS LES L G+P++AWPL+A+Q+M
Sbjct: 184 LERTKGQGLVVPLWAPQVQVLGHRSVGGFLSHCGWNSTLESVLQGVPLIAWPLFAEQRMN 363
Query: 400 AALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKE 459
A L +GLKV L P+ NE G+V RG +A+VIK ++ GEEG E+++RM L++ A +A+KE
Sbjct: 364 AILLCEGLKVGLWPRVNENGLVERGEIAKVIKCLMGGEEGGELRRRMTELKEAATNAIKE 543
Query: 460 DGSSTRTITQLALKWKRSA 478
+GSST+ + Q LKWK+ A
Sbjct: 544 NGSSTKALAQAVLKWKKLA 600
>TC233949 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (34%)
Length = 638
Score = 219 bits (557), Expect = 3e-57
Identities = 105/189 (55%), Positives = 136/189 (71%), Gaps = 1/189 (0%)
Frame = +2
Query: 288 TLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHE-DPLEFLPNGFLERTKGQ 346
TL+ +Q NELA GLELS K LWV+R P L LS + DPL F P GFLERTK Q
Sbjct: 2 TLSQEQINELACGLELSNYKXLWVVRAPSSLASDAYLSAQKDVDPLHFFPCGFLERTKEQ 181
Query: 347 GFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDG 406
G VVP WA QI++L H ++GGFL HCGWNS+LE L G+P + WPL+A+Q+MIA L +G
Sbjct: 182 GMVVPSWAPQIQVLSHSSVGGFLTHCGWNSILEGVLKGVPFITWPLFAEQRMIAVLLCEG 361
Query: 407 LKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRT 466
LKV +RP+ +E G+V R + +VIK ++ GEEG ++ RM L++ A +ALKEDGSST+T
Sbjct: 362 LKVGVRPRVSENGLVEREEIVKVIKCLMEGEEGGKMSGRMNELKEAATNALKEDGSSTKT 541
Query: 467 ITQLALKWK 475
++ LALKWK
Sbjct: 542 LSLLALKWK 568
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 213 bits (543), Expect = 1e-55
Identities = 131/352 (37%), Positives = 196/352 (55%), Gaps = 8/352 (2%)
Frame = +3
Query: 134 NVLSYVYFSSTAMLLSLCLYPSTFDKTISCET-------VEIPGCMLIHTTDLPDPVQNR 186
NV +Y Y++S A L+L LY T T+ E ++IPG I D P+ ++
Sbjct: 99 NVPTYFYYTSGASTLALLLYYPTIHPTLIEEKDTDQPLQIQIPGLSTITADDFPNECKDP 278
Query: 187 FRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPVGPLV 246
S +VFL+ + G+I+N+F +E E RAL E+ +T P ++ VGP++
Sbjct: 279 L--SYACQVFLQIAETMMGGAGIIVNTFEAIEEEAIRALSED----ATVPPPLFCVGPVI 440
Query: 247 QEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGV 306
E E + WL+ Q SV+ + FGS G + Q E+A+GLE S
Sbjct: 441 SAPYGE---------EDKGCLSWLNLQPSQSVVLLCFGSMGRFSRAQLKEIAIGLEKSEQ 593
Query: 307 KFLWVIRPPQKLEMIGDLSVGHEDPL-EFLPNGFLERTKGQGFVVPYWADQIEILGHGAI 365
+FLWV+R E+ G E L E LP GFLERTK +G VV WA Q IL H ++
Sbjct: 594 RFLWVVRT----ELGGADDSAEELSLDELLPEGFLERTKEKGMVVRDWAPQAAILSHDSV 761
Query: 366 GGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRGV 425
GGF+ HCGWNS+LE+ G+P+VAWPLYA+QKM + +KVAL K N+ G V
Sbjct: 762 GGFVTHCGWNSVLEAVCEGVPMVAWPLYAEQKMNRMVMVKEMKVALAVKENKDGFVSSTE 941
Query: 426 VAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWKRS 477
+ + ++ ++ ++GKEI+QR+ +++ A +A+ E G+S ++ +LA WK+S
Sbjct: 942 LGDRVRELMESDKGKEIRQRIFKMKMSAAEAMAEGGTSRASLDKLAKLWKQS 1097
>TC204806 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (41%)
Length = 1014
Score = 196 bits (497), Expect = 3e-50
Identities = 95/192 (49%), Positives = 135/192 (69%), Gaps = 4/192 (2%)
Frame = +1
Query: 287 GTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSV----GHEDPLEFLPNGFLER 342
GTL+ Q NELALGLE S +FLWV++ P E I + + DPL+FLP GF+ER
Sbjct: 4 GTLSSAQINELALGLEKSEQRFLWVVKSPN--EEIANATYFSAESQADPLQFLPEGFVER 177
Query: 343 TKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAAL 402
TKG+GF+V WA Q ++LGH + GGFL HCGWNS+LES ++G+P +AWPL+A+Q+ A +
Sbjct: 178 TKGRGFLVQSWAPQPQVLGHPSTGGFLTHCGWNSILESVVNGVPFIAWPLFAEQRTNAFM 357
Query: 403 FSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGS 462
+ +KVALRP E G+V R +A ++K ++ GE+GK+++ R+K +++ A AL + GS
Sbjct: 358 LTHDVKVALRPNVAESGLVERQEIASLVKCLMEGEQGKKLRYRIKDIKEAAAKALAQHGS 537
Query: 463 STRTITQLALKW 474
ST I+ LALKW
Sbjct: 538 STTNISNLALKW 573
>TC212628 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (36%)
Length = 704
Score = 168 bits (426), Expect = 4e-42
Identities = 96/219 (43%), Positives = 132/219 (59%), Gaps = 6/219 (2%)
Frame = +1
Query: 1 MEVTTTTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENL 60
+ + TTHIAIVS P Y+H I+EF KRL HQ+ H+TCI P++ KA + L
Sbjct: 61 VSMAKTTHIAIVSSPGYTHLVPIIEFSKRLIKHHQNFHVTCIVPSLGPPPESSKAYLKTL 240
Query: 61 PPNIDSMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVD 120
P NID++ LPP++ + +P+ H IL+ TIT SLPSI+ ALK+L CS LT++VVD
Sbjct: 241 PSNIDTILLPPISKEQLPQGVHPAILIQLTITLSLPSIHEALKSL-CS--KAPLTALVVD 411
Query: 121 VLITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTIS------CETVEIPGCMLI 174
V Q L A E N LSY YF S+AM+LSL ++ ++ +S E + +PGC+ +
Sbjct: 412 VFAFQALEYAKEFNALSYFYFPSSAMILSLLMHAPKLNEEVSGEYKDLTEPIRLPGCVPV 591
Query: 175 HTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINS 213
DLPDP Q+ R SE Y LE ADG++IN+
Sbjct: 592 MGVDLPDPAQD--RSSEIYNHXLERAXAMVTADGILINT 702
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 164 bits (415), Expect = 8e-41
Identities = 133/460 (28%), Positives = 210/460 (44%), Gaps = 15/460 (3%)
Frame = +1
Query: 7 THIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVK-SSCNDVKALFENLPPNID 65
T + ++ P H +L + + + +H ++ ++ D + N+ +
Sbjct: 34 TQVVLIPFPAQGHLNQLLHLARHIFSHNIPVHYVGTATHIRQATLRDHNSNISNIIIHFH 213
Query: 66 SMFLPP-VNLDDMPEKPHIPILVHATITSSLPS-----IYNALKTLHCSSNSIGLTSIVV 119
+ +PP V+ P H + S + N L++L + + ++
Sbjct: 214 AFEVPPFVSPPPNPNNEETDFPSHLLPSFKASSHLREPVRNLLQSLSSQAKRV---IVIH 384
Query: 120 DVLITQVLPMANEL-NVLSYVYFSSTAMLLSLCLYPSTFDKTIS--CETVEIP---GCML 173
D L+ V A + NV +Y + S+ A L + + + EIP GC
Sbjct: 385 DSLMASVAQDATNMPNVENYTFHSTCAFTTFLYYWEVMGRPPVEGFFQATEIPSMGGCF- 561
Query: 174 IHTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSS 233
P Q +EEY+ F + N DG I N+ +E L E G S
Sbjct: 562 --------PPQFIHFITEEYE-FHQFN------DGNIYNTSRAIEGPYIEFL-ERIGGSK 693
Query: 234 TSIPHVYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQ 293
+ + P PL E + + H + WLDKQE NSV+YVSFG+ + T Q
Sbjct: 694 KRLWALGPFNPLTIE--------KKDPKTRHICIEWLDKQEANSVMYVSFGTTTSFTVAQ 849
Query: 294 FNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYW 353
F ++A+GLE S KF+WV+R K G++ G E LPNGF ER +G G ++ W
Sbjct: 850 FEQIAIGLEQSKQKFIWVLRDADK----GNIFDGSEAERYELPNGFEERVEGIGLLIRDW 1017
Query: 354 ADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRP 413
A Q+EIL H + GGF+ HCGWNS LES G+PI AWP+++DQ + L ++ LKV
Sbjct: 1018APQLEILSHTSTGGFMSHCGWNSCLESITMGVPIAAWPMHSDQPRNSVLITEVLKVGFVV 1197
Query: 414 K--PNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQD 451
K +V VV ++ ++ +EG E++ R RL++
Sbjct: 1198KDWAQRNALVSASVVENAVRRLMETKEGDEMRDRAVRLKN 1317
>NP163422 UDP-glycose:flavonoid glycosyltransferase
Length = 731
Score = 163 bits (413), Expect = 1e-40
Identities = 97/253 (38%), Positives = 138/253 (54%), Gaps = 6/253 (2%)
Frame = +1
Query: 129 MANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCET------VEIPGCMLIHTTDLPDP 182
+ N L + +Y Y++S A L++ LY + F + + VEIPG IHT D+P+
Sbjct: 19 VTNTLQIPTYFYYTSGASTLAVFLYQTIFHENYTKSLKDLNMHVEIPGLPKIHTDDMPET 198
Query: 183 VQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPV 242
VQ+R + E Y+VF++ +DGVI+N+ +E A E G + P V+ +
Sbjct: 199 VQDRAK--EVYQVFIDIATCMRDSDGVIVNTCEAMEERVVEAFSE--GLMEGTTPKVFCI 366
Query: 243 GPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLE 302
GP++ SC ++ E + WLD Q +SVL++SFGS G + Q E+A+GLE
Sbjct: 367 GPVIASASCRRDDN--------ECLSWLDSQPSHSVLFLSFGSMGRFSRTQLGEIAIGLE 522
Query: 303 LSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGH 362
S +FLWV+R E SV E LP GFLERTK +G VV WA Q IL H
Sbjct: 523 KSEQRFLWVVRS----EFENGDSVEPPSLDELLPEGFLERTKEKGMVVRDWAPQAAILSH 690
Query: 363 GAIGGFLCHCGWN 375
++GGF+ HCGWN
Sbjct: 691 DSVGGFVTHCGWN 729
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 161 bits (407), Expect = 7e-40
Identities = 129/471 (27%), Positives = 208/471 (43%), Gaps = 8/471 (1%)
Frame = +1
Query: 5 TTTHIAIVSVPLYSHTRTILEFCKRLAHLHQDIH----ITCINPTVKSSCNDVKALFENL 60
T ++ P H +L + + + +H +T I N + + +
Sbjct: 34 TQVMAVLIPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFHA 213
Query: 61 PPNIDSMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVD 120
+ S PP N ++ E+ P + + +S K LH S+ ++ D
Sbjct: 214 F-EVPSFVSPPPNPNN--EETDFPAHLLPSFEASSHLREPVRKLLHSLSSQAKRVIVIHD 384
Query: 121 VLITQVLPMANEL-NVLSYVYFSSTAMLLSLCLYPSTFDKTISCETV-EIPGCMLIHTTD 178
++ V A + NV +Y + S+ ++ + + V EIP TTD
Sbjct: 385 SVMASVAQDATNMPNVENYTFHSTCTFGTAVFYWDKMGRPLVDGMLVPEIPSMEGCFTTD 564
Query: 179 LPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPH 238
N +++ +GN I N+ +E ++ + T +
Sbjct: 565 F----MNFMIAQRDFRKVNDGN---------IYNTSRAIEGAYIEWME--RFTGGKKLWA 699
Query: 239 VYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELA 298
+ P PL E + + E H + WLDKQ+ NSVLYVSFG+ T +Q ++A
Sbjct: 700 LGPFNPLAFE--------KKDSKERHFCLEWLDKQDPNSVLYVSFGTTTTFKEEQIKKIA 855
Query: 299 LGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIE 358
GLE S KF+WV+R K GD+ G E N F ER +G G VV WA Q+E
Sbjct: 856 TGLEQSKQKFIWVLRDADK----GDIFDGSEAKWNEFSNEFEERVEGMGLVVRDWAPQLE 1023
Query: 359 ILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPK--PN 416
IL H + GGF+ HCGWNS LES G+PI AWP+++DQ + L ++ LK+ L K
Sbjct: 1024ILSHTSTGGFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGLVVKNWAQ 1203
Query: 417 EKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTI 467
+V V ++ ++ +EG ++++R RL++V ++ E G S I
Sbjct: 1204RNALVSASNVENAVRRLMETKEGDDMRERAVRLKNVIHRSMDEGGVSRMEI 1356
>BG156821 similar to SP|Q9AR73|HQGT Hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase). [Serpentwood
Devilpepper], partial (27%)
Length = 465
Score = 160 bits (406), Expect = 9e-40
Identities = 84/166 (50%), Positives = 110/166 (65%), Gaps = 6/166 (3%)
Frame = +2
Query: 232 SSTSIPHVYPVGPLV--QEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTL 289
SS P VY VGPLV + G ++ E +RWLD+Q + SVL+VSFGS GTL
Sbjct: 5 SSPGRPPVYAVGPLVRMEPGPADS-----------ECLRWLDEQPRGSVLFVSFGSGGTL 151
Query: 290 THDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSV----GHEDPLEFLPNGFLERTKG 345
+ Q NELALGLE S +FLWV++ P + I + + HEDPL+FLP GF+ERTKG
Sbjct: 152 SSAQINELALGLENSQQRFLWVVKSPN--DAIANATYFNAESHEDPLQFLPEGFVERTKG 325
Query: 346 QGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWP 391
+G +V WA Q + L H + GGFL HCGWNS+LES ++G+P++AWP
Sbjct: 326 RGXLVKSWAPQPQGLAHQSTGGFLSHCGWNSILESVVNGVPLIAWP 463
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 159 bits (403), Expect = 2e-39
Identities = 130/478 (27%), Positives = 220/478 (45%), Gaps = 19/478 (3%)
Frame = +2
Query: 11 IVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSMFLP 70
++ P H +++F KRLA + + T +S N E + D
Sbjct: 5 VLPYPAQGHINPLVQFAKRLASKGVKATVATTHYTA-NSINAPNITVEAISDGFDQAGFA 181
Query: 71 PVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVLPMA 130
N ++ + + + T+ ++ ++ H + S T IV D VL +A
Sbjct: 182 QTN-------NNVQLFLASFRTNGSRTLSELIRK-HQQTPSPS-TCIVYDSFFPWVLDVA 334
Query: 131 NELNVLSYVYFSSTAMLLSLC--LYPSTFDKTISCE--TVEIPGCMLIHTTDLPDPVQNR 186
+ + +F+++A + ++ L+ + E + +PG + + LP V
Sbjct: 335 KQHGIYGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLRVPGLPPLDSRALPSFV--- 505
Query: 187 FRCSEEYKVF----LEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYP- 241
R E Y + L AD + +N+F LE E + L E ++P
Sbjct: 506 -RFPESYPAYMAMKLSQFSNLNNADWMFVNTFEALESEVLKGLTE-----------LFPA 649
Query: 242 --VGPLVQEGSCETHESQGNEGE--------THEYMRWLDKQEQNSVLYVSFGSAGTLTH 291
+GP+V G + +G++G T E WL+ + SV+Y+SFGS +LT
Sbjct: 650 KMIGPMVPSGYLDGR-IKGDKGYGASLWKPLTEECSNWLESKPPQSVVYISFGSMVSLTE 826
Query: 292 DQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVP 351
+Q E+A GL+ SGV FLWV+R + + LP G+ E K +G +V
Sbjct: 827 EQMEEVAWGLKESGVSFLWVLRESEHGK---------------LPLGYRESVKDKGLIVT 961
Query: 352 YWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVAL 411
W +Q+E+L H A G F+ HCGWNS LES G+P+V P +ADQ A + +V +
Sbjct: 962 -WCNQLELLAHQATGCFVTHCGWNSTLESLSLGVPVVCLPQWADQLPDAKFLDEIWEVGV 1138
Query: 412 RPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQ 469
PK +EKGIV + + +K+++ G+ +EI++ + + +A +A+ E GSS + I Q
Sbjct: 1139WPKEDEKGIVRKQEFVQSLKDVMEGQRSQEIRRNANKWKKLAREAVGEGGSSDKHINQ 1312
>TC223096 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (22%)
Length = 677
Score = 157 bits (398), Expect = 8e-39
Identities = 70/127 (55%), Positives = 98/127 (77%)
Frame = +1
Query: 349 VVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLK 408
VVP WA QI++L H ++GGFL HCGWNS+LES L G+P + WPL+A+QKM A L S+GLK
Sbjct: 4 VVPSWAPQIQVLSHSSVGGFLTHCGWNSILESVLKGVPFITWPLFAEQKMNAVLLSEGLK 183
Query: 409 VALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTIT 468
V +RP+ +E G+V R + +VIK ++ GEEG ++++RM L++ A +ALKEDGSST+ ++
Sbjct: 184 VGVRPRVSENGLVERVEIVDVIKCLMEGEEGAKMRERMNELKEDATNALKEDGSSTKALS 363
Query: 469 QLALKWK 475
QL L WK
Sbjct: 364 QLPLYWK 384
>TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferase-like
protein , partial (36%)
Length = 1202
Score = 155 bits (392), Expect = 4e-38
Identities = 96/275 (34%), Positives = 152/275 (54%), Gaps = 10/275 (3%)
Frame = +2
Query: 206 ADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPVGPLVQEGSC-ETHESQGNEG--E 262
+DG I N+ ++EP + L+ + +P V+PVGPL+ S ++ G E
Sbjct: 314 SDGWICNTVQEIEPLGLQLLR-----NYLQLP-VWPVGPLLPPASLMDSKHRAGKESGIA 475
Query: 263 THEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIG 322
M+WLD ++++SVLY+SFGS T+T Q LA GLE SG F+W+IRPP ++ G
Sbjct: 476 LDACMQWLDSKDESSVLYISFGSQNTITASQMMALAEGLEESGRSFIWIIRPPFGFDING 655
Query: 323 DLSVGHEDPLEFLPNGFLERTKG--QGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLES 380
+ E+LP GF ER + +G +V W Q+EIL H + G FL HCGWNS+LES
Sbjct: 656 EFIA------EWLPKGFEERMRDTKRGLLVNKWGPQLEILSHSSTGAFLSHCGWNSVLES 817
Query: 381 TLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGE-EG 439
+G+P++ WPL A+Q + + + VA+ + ++ V +VI+ + E +G
Sbjct: 818 LSYGVPMIGWPLAAEQTYNLKMLVEEMGVAIELTQTVETVISGKQVKKVIEIAMEQEGKG 997
Query: 440 KEIQQRMK----RLQDVAIDALKEDGSSTRTITQL 470
KE++++ +++ + KE GSS R + L
Sbjct: 998 KEMKEKANEIAAHMREAITEKGKEKGSSVRAMDDL 1102
>TC220035 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (29%)
Length = 706
Score = 148 bits (374), Expect = 5e-36
Identities = 82/190 (43%), Positives = 122/190 (64%), Gaps = 11/190 (5%)
Frame = +3
Query: 293 QFNELALGLELSGVKFLWVIRPPQKLEMIGDL------SVGHEDPLEFLPNGFLERTKGQ 346
Q ELA GLELS +F+WV+R P +E D S G ++ ++LP GF+ RT+
Sbjct: 3 QMTELAWGLELSEWRFVWVVRAP--MEGTADAAFFTTGSDGVDEVAKYLPEGFVSRTRKV 176
Query: 347 GFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDG 406
G +VP WA Q+ IL H +IGGFL HCGW S LES +GIP++AWPLYA+Q+M A L ++
Sbjct: 177 GLLVPEWAQQVTILKHRSIGGFLSHCGWGSTLESVTNGIPLIAWPLYAEQRMNATLLAEE 356
Query: 407 LKVALRPK--PNEKGIVGRGVVAEVIKNILVGEEGKE---IQQRMKRLQDVAIDALKEDG 461
L +A+R P +K +V R +A +++ +L G+E + I++R+K +Q A++AL E G
Sbjct: 357 LGLAVRTTVLPTKK-VVRREEIARMVREVLQGDENVKSNGIRERVKEVQRSAVNALSEGG 533
Query: 462 SSTRTITQLA 471
SS ++ +A
Sbjct: 534 SSYVALSHVA 563
>TC210204 similar to UP|Q9SY84 (Q9SY84) F14N23.30, partial (21%)
Length = 782
Score = 145 bits (367), Expect = 3e-35
Identities = 77/201 (38%), Positives = 121/201 (59%), Gaps = 3/201 (1%)
Frame = +3
Query: 273 QEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPL 332
+E++SVLY +FGS ++ +Q E+A GLE S V FLWVIR +
Sbjct: 33 EEKSSVLYAAFGSQAEISREQLEEIAKGLEESKVSFLWVIRKEEW--------------- 167
Query: 333 EFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPL 392
LP+G+ ER K +G V+ W DQ EIL H ++ GFL HCGWNS++ES G+PIV WP+
Sbjct: 168 -GLPDGYEERVKDRGIVIREWVDQREILMHESVEGFLSHCGWNSVMESVTAGVPIVGWPI 344
Query: 393 YADQKMIAALFSDGLKVALRPKP---NEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRL 449
A+Q + A + + +KV LR + + +G V R + + +K ++ G +GK+++++++ L
Sbjct: 345 MAEQFLNARMVEEEVKVGLRVETCDGSVRGFVKREGLKKTVKEVMEGVKGKKLREKVREL 524
Query: 450 QDVAIDALKEDGSSTRTITQL 470
++A A +E GSS T+ L
Sbjct: 525 AEMAKLATQEGGSSCSTLNSL 587
>TC231164 weakly similar to UP|UFO2_MANES (Q40285) Flavonol
3-O-glucosyltransferase 2 (UDP-glucose flavonoid
3-O-glucosyltransferase 2) (Fragment) , partial (47%)
Length = 901
Score = 145 bits (366), Expect = 4e-35
Identities = 82/207 (39%), Positives = 120/207 (57%), Gaps = 3/207 (1%)
Frame = +2
Query: 267 MRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSV 326
M WLD +SV+ FGS G+L Q E+A GLE++ V+FLW +R P K ++
Sbjct: 14 MEWLDXXXVSSVVXXCFGSMGSLXXXQVEEIATGLEMANVRFLWALREPPKAQLEDPRD- 190
Query: 327 GHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIP 386
+ +P + LP+GFLERT G V W Q +L H A+GGF+ HCGWNS+LES HG+P
Sbjct: 191 -YXNPKDVLPDGFLERTAEMGLVCG-WVPQAVVLAHKAVGGFVSHCGWNSILESLWHGVP 364
Query: 387 IVAWPLYADQKMIA--ALFSDGLKVALRPKPNEKG-IVGRGVVAEVIKNILVGEEGKEIQ 443
I WP+YA+Q+M A + GL V +R G +V V +++++ G + EIQ
Sbjct: 365 IATWPVYAEQQMNAFQMVRELGLAVEIRVDYRVGGDLVRAEEVLNGVRSLMKGAD--EIQ 538
Query: 444 QRMKRLQDVAIDALKEDGSSTRTITQL 470
+++K + D+ AL E+ SS + L
Sbjct: 539 KKVKEMSDICRSALMENRSSYNNLVFL 619
>BE022618 weakly similar to GP|19911209|db glucosyltransferase-13 {Vigna
angularis}, partial (19%)
Length = 400
Score = 144 bits (363), Expect = 9e-35
Identities = 67/126 (53%), Positives = 95/126 (75%)
Frame = +1
Query: 353 WADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALR 412
WA Q+++L H ++GGFL HCGWNS LES G+PI+ WPL+A+Q+M A + +DGLKVALR
Sbjct: 10 WAPQVQVLSHNSVGGFLSHCGWNSTLESVQEGVPIITWPLFAEQRMNAVMLTDGLKVALR 189
Query: 413 PKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLAL 472
K NE GIV + +A VIK ++ GEEGK +++RM L+D+ +ALK DG T+T++QLA
Sbjct: 190 TKFNEDGIVEKEEIARVIKCLMEGEEGKGMRERMMNLKDLFGNALK-DGFLTQTLSQLAR 366
Query: 473 KWKRSA 478
+W+ S+
Sbjct: 367 RWENSS 384
>TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, partial
(60%)
Length = 1281
Score = 144 bits (362), Expect = 1e-34
Identities = 95/275 (34%), Positives = 140/275 (50%), Gaps = 19/275 (6%)
Frame = +1
Query: 208 GVIINSFFDLEPETFRALQENKGTSSTSIPHVYPVGPLVQEGSCETHESQGNEGETHEYM 267
G +NSF DLEP ++ G + I PV + +T + + + +
Sbjct: 88 GTFVNSFHDLEPAYAEQVKNKWGKKAWIIG---PVSLCNRTAEDKTERGKLPTIDEEKCL 258
Query: 268 RWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVG 327
WL+ ++ NSVLYVSFGS L +Q E+A GLE S F+WV+R
Sbjct: 259 NWLNSKKPNSVLYVSFGSLLRLPSEQLKEIACGLEASEQSFIWVVRNI------------ 402
Query: 328 HEDPLE--------FLPNGFLERTK--GQGFVVPYWADQIEILGHGAIGGFLCHCGWNSM 377
H +P E FLP GF +R K +G V+ WA Q+ IL H AI GF+ HCGWNS
Sbjct: 403 HNNPSENKENGNGNFLPEGFEQRMKEKDKGLVLRGWAPQLLILEHVAIKGFMTHCGWNST 582
Query: 378 LESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNE--------KGIVGRGVVAEV 429
LES G+P++ WPL A+Q L ++ LK+ ++ E K +VGR V
Sbjct: 583 LESVCAGVPMITWPLSAEQFSNEKLITEVLKIGVQVGSREWLSWNSEWKDLVGREKVESA 762
Query: 430 IKNILV-GEEGKEIQQRMKRLQDVAIDALKEDGSS 463
++ ++V EE +E+ R+K + + A A++E G+S
Sbjct: 763 VRKLMVESEEAEEMTTRVKDIAEKAKRAVEEGGTS 867
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,822,105
Number of Sequences: 63676
Number of extensions: 345915
Number of successful extensions: 2091
Number of sequences better than 10.0: 181
Number of HSP's better than 10.0 without gapping: 1958
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1968
length of query: 479
length of database: 12,639,632
effective HSP length: 101
effective length of query: 378
effective length of database: 6,208,356
effective search space: 2346758568
effective search space used: 2346758568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0135.13


