

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.5
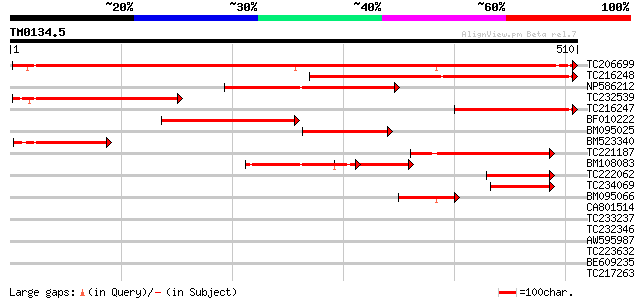
(510 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206699 UP|Q7XA53 (Q7XA53) Sucrose transporter, complete 710 0.0
TC216248 similar to UP|O04077 (O04077) Sucrose transport protein... 371 e-103
NP586212 hypothetical protein [Glycine max] 235 3e-62
TC232539 similar to UP|Q7XA53 (Q7XA53) Sucrose transporter, part... 230 1e-60
TC216247 similar to UP|O04077 (O04077) Sucrose transport protein... 180 1e-45
BF010222 similar to GP|28172870|em sucrose transporter 4 protein... 140 2e-33
BM095025 136 2e-32
BM523340 126 2e-29
TC221187 similar to UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 pro... 122 4e-28
BM108083 111 9e-25
TC222062 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, pa... 88 1e-17
TC234069 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, pa... 82 6e-16
BM095066 56 4e-08
CA801514 homologue to GP|7649151|gb|A sucrose transport protein ... 33 0.40
TC233237 33 0.40
TC232346 similar to UP|Q96469 (Q96469) Calcium dependent protein... 32 0.52
AW595987 30 2.0
TC223632 30 2.0
BE609235 weakly similar to SP|P08926|RUBA RuBisCO subunit bindin... 30 3.4
TC217263 similar to UP|Q949X7 (Q949X7) Diaminopimelate decarboxy... 30 3.4
>TC206699 UP|Q7XA53 (Q7XA53) Sucrose transporter, complete
Length = 2053
Score = 710 bits (1833), Expect = 0.0
Identities = 358/519 (68%), Positives = 422/519 (80%), Gaps = 11/519 (2%)
Frame = +1
Query: 3 SPTKSNHLDTKG---SSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQ 59
S TK NH + SL +E+ P + SPLRK+I VASIAAG+QFGWALQLSLLTPYVQ
Sbjct: 28 SSTKQNHNNNNTLTKPSLHVESP-PLEPSPLRKIIVVASIAAGVQFGWALQLSLLTPYVQ 204
Query: 60 TLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIG 119
LG+PH WA++IWLCGP+SG+LVQPIVGY+SDR T RFGRRRPFI AG+ AVAI+VFLIG
Sbjct: 205 LLGIPHTWAAYIWLCGPISGMLVQPIVGYHSDRCTSRFGRRRPFIAAGSFAVAIAVFLIG 384
Query: 120 YAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRT 179
YAAD+GHS GDDL+KK RPRA+ IFV GFWILDVANNMLQGPCRA LGDL AG+ +KTR
Sbjct: 385 YAADLGHSFGDDLSKKVRPRAIGIFVVGFWILDVANNMLQGPCRALLGDLCAGNHQKTRN 564
Query: 180 AMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSI 239
A FFSFFMAVGNVLGYAAGAYS L+ +FPFT T AC +CANLKSCFF SI LL L+
Sbjct: 565 ANAFFSFFMAVGNVLGYAAGAYSKLYHVFPFTKTTACDVYCANLKSCFFLSIALLTTLAT 744
Query: 240 AALIYVEDTPLTKKPA---ADVDSPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPF 296
AAL+YV++ PL+ + A +D + + CFG LFGAF+ELK+PMWIL+LVT +NW+AWFPF
Sbjct: 745 AALVYVKEVPLSPEKAVIDSDDNGGMPCFGQLFGAFRELKRPMWILLLVTCLNWIAWFPF 924
Query: 297 FLFDTDWMGREVYGGVVGE-KAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKN 355
LFDTDWMGREVY G VGE KAYD GVRAG+LGLM+N+VVL SL VE L R +GGVK
Sbjct: 925 LLFDTDWMGREVYEGTVGEGKAYDRGVRAGALGLMLNSVVLGATSLGVEVLARGVGGVKR 1104
Query: 356 LWGIVNFILAICMAMTVLITKTAEHDR----LISGGATVGAPSSGVKAGAIAFFGVMGIP 411
LWGIVNF+LA+C+AMTVL+TK A+H R L + P + VKAGA+A F ++GIP
Sbjct: 1105LWGIVNFLLAVCLAMTVLVTKMAQHSRQYTLLPNAHQEPLPPPAAVKAGALALFSLLGIP 1284
Query: 412 LAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAF 471
LAI +S+PFALASI+SS +GAGQGLSLGVLNLAIV+PQM+VS +SGPWDALFGGGNLPAF
Sbjct: 1285LAITYSIPFALASIFSSTSGAGQGLSLGVLNLAIVIPQMVVSVISGPWDALFGGGNLPAF 1464
Query: 472 VVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH 510
VVGAV AA S I++I+LLP+P P D+AKA++ GGGFH
Sbjct: 1465VVGAVAAAASGILSIILLPSP-PPDLAKAAT-AAGGGFH 1575
>TC216248 similar to UP|O04077 (O04077) Sucrose transport protein, partial
(41%)
Length = 1234
Score = 371 bits (953), Expect = e-103
Identities = 187/241 (77%), Positives = 207/241 (85%)
Frame = +1
Query: 270 GAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVGEKAYDDGVRAGSLGL 329
GA KELK+PMW+LMLVTAVNWV WFP+FLFDTDWMGREVYGG GE AY GVR GSLGL
Sbjct: 1 GALKELKRPMWMLMLVTAVNWVGWFPYFLFDTDWMGREVYGGTAGEDAYAKGVRVGSLGL 180
Query: 330 MINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDRLISGGAT 389
M+NAVVL FMSLAVEPLG+++GGVK LW IVNFILAI MTV+ITK AEH R ++ A
Sbjct: 181 MVNAVVLGFMSLAVEPLGKMVGGVKRLWAIVNFILAIGFGMTVVITKVAEHQRRMNPAA- 357
Query: 390 VGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQ 449
VG PS GV G++ FFGV+G+PLAI FSVPFALASIY SA+GAGQGLSLGVLNLAIVVPQ
Sbjct: 358 VGHPSEGVVVGSMVFFGVLGVPLAITFSVPFALASIYCSASGAGQGLSLGVLNLAIVVPQ 537
Query: 450 MLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGF 509
M+VS LSGPWD+LFGGGNLPAF+VGA AA+SAIMAIVLLPTPKPAD AKASS+ GGF
Sbjct: 538 MVVSALSGPWDSLFGGGNLPAFMVGAAAAALSAIMAIVLLPTPKPADEAKASSM-MAGGF 714
Query: 510 H 510
H
Sbjct: 715 H 717
>NP586212 hypothetical protein [Glycine max]
Length = 470
Score = 235 bits (600), Expect = 3e-62
Identities = 113/157 (71%), Positives = 128/157 (80%)
Frame = +2
Query: 194 LGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKK 253
LGYAAG+Y GLHK+FPFT T AC FCANLKSCFFFSILLLL L+ AL+YV+D + +
Sbjct: 2 LGYAAGSYKGLHKMFPFTETKACDVFCANLKSCFFFSILLLLFLATVALLYVKDKQVEAR 181
Query: 254 PAADVDSPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVV 313
D P SCF LF A KELK+PMW+LMLVTAVNWV WFP+FLFDTDWMGREVYGG V
Sbjct: 182 ALDDATQP-SCFFQLFSALKELKRPMWMLMLVTAVNWVGWFPYFLFDTDWMGREVYGGQV 358
Query: 314 GEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRIL 350
GE AY +GVR GSLGLM+NAVVL FMSLAVEPLG+++
Sbjct: 359 GEDAYANGVRVGSLGLMVNAVVLGFMSLAVEPLGKMV 469
>TC232539 similar to UP|Q7XA53 (Q7XA53) Sucrose transporter, partial (25%)
Length = 591
Score = 230 bits (586), Expect = 1e-60
Identities = 119/157 (75%), Positives = 132/157 (83%), Gaps = 4/157 (2%)
Frame = +1
Query: 3 SPTKSNHLDTKGSS----LQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYV 58
SPTK N TK S+ L +EA P Q+SPLRKM AVASIAAGIQFGWALQLSLLTPYV
Sbjct: 112 SPTKPND-PTKPSNTSTTLSLEAG-PAQASPLRKMFAVASIAAGIQFGWALQLSLLTPYV 285
Query: 59 QTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLI 118
Q LGVPH ASFIWLCGP+SGL+VQPIVGYYSD T RFGRRRPFIL GA+AVA++VFLI
Sbjct: 286 QLLGVPHAAASFIWLCGPISGLVVQPIVGYYSDHCTSRFGRRRPFILGGALAVAVAVFLI 465
Query: 119 GYAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDVAN 155
GYAADIG++ GDD++K TRPRAV +FV GFWILDVAN
Sbjct: 466 GYAADIGYAAGDDISKTTRPRAVGVFVIGFWILDVAN 576
>TC216247 similar to UP|O04077 (O04077) Sucrose transport protein, partial
(17%)
Length = 930
Score = 180 bits (457), Expect = 1e-45
Identities = 92/110 (83%), Positives = 101/110 (91%)
Frame = +2
Query: 401 AIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWD 460
++ FFGV+G+PLAI FSVPFALASIY SA+GAGQGLSLGVLNLAIVVPQM+VSTLSGPWD
Sbjct: 2 SMVFFGVLGVPLAITFSVPFALASIYCSASGAGQGLSLGVLNLAIVVPQMVVSTLSGPWD 181
Query: 461 ALFGGGNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH 510
ALFGGGNLPAF+VGA AA+SAIMAIVLLPTPKPAD AKASS+ GGFH
Sbjct: 182 ALFGGGNLPAFMVGAAAAALSAIMAIVLLPTPKPADEAKASSM-MAGGFH 328
>BF010222 similar to GP|28172870|em sucrose transporter 4 protein {Lotus
japonicus}, partial (26%)
Length = 414
Score = 140 bits (352), Expect = 2e-33
Identities = 61/124 (49%), Positives = 85/124 (68%)
Frame = +1
Query: 137 RPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGY 196
RP A+ +F+ GFWILDVANN+ QGPCRA LGDL + D ++TR A ++S FMA+GN+LGY
Sbjct: 13 RPAAITVFIVGFWILDVANNVTQGPCRALLGDLTSKDPRRTRVANAYYSLFMAIGNILGY 192
Query: 197 AAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAA 256
A G+YSG +KIF F ++ AC CANLKS FF I + + + +++ + PL AA
Sbjct: 193 ATGSYSGWYKIFTFALSPACTISCANLKSAFFLDIAFIAVTTYISIMAAHEVPLNSSEAA 372
Query: 257 DVDS 260
++
Sbjct: 373 HAEA 384
>BM095025
Length = 335
Score = 136 bits (343), Expect = 2e-32
Identities = 63/82 (76%), Positives = 71/82 (85%), Gaps = 1/82 (1%)
Frame = +3
Query: 264 CFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVGE-KAYDDGV 322
CFG LFGAF+ELK+PMWIL+LVT +NW+AWFPF LFDTDWMGREVY G VGE KAYD GV
Sbjct: 90 CFGQLFGAFRELKRPMWILLLVTCLNWIAWFPFLLFDTDWMGREVYEGTVGEGKAYDRGV 269
Query: 323 RAGSLGLMINAVVLAFMSLAVE 344
RAG+LGLM+N+VVL SL VE
Sbjct: 270 RAGALGLMLNSVVLGATSLGVE 335
>BM523340
Length = 429
Score = 126 bits (317), Expect = 2e-29
Identities = 65/88 (73%), Positives = 73/88 (82%)
Frame = +2
Query: 4 PTKSNHLDTKGSSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGV 63
PTK ++ S+L +E P ++SPLRKM AVASIAAGIQFGWALQLSLLTPYVQ LGV
Sbjct: 170 PTKPSNTS---STLSLEGG-PGEASPLRKMFAVASIAAGIQFGWALQLSLLTPYVQLLGV 337
Query: 64 PHIWASFIWLCGPVSGLLVQPIVGYYSD 91
PH ASFIWLCGP+SGL+VQPIVGYYSD
Sbjct: 338 PHAAASFIWLCGPISGLVVQPIVGYYSD 421
>TC221187 similar to UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 protein,
partial (26%)
Length = 977
Score = 122 bits (306), Expect = 4e-28
Identities = 60/130 (46%), Positives = 85/130 (65%)
Frame = +2
Query: 361 NFILAICMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPF 420
N ++ +C +++T A + G P +G+ A+ F ++G PLAI +SVP+
Sbjct: 5 NIMMTVCFLAMLVVTYVANN----MGYIGKDLPPTGIVIAALIIFTILGFPLAITYSVPY 172
Query: 421 ALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAV 480
AL S + + G GQGLS+GVLNLAIVVPQ++VS SGPWD LFGGGN PAF V AV A +
Sbjct: 173 ALISTHIESLGLGQGLSMGVLNLAIVVPQIIVSLGSGPWDQLFGGGNSPAFAVAAVSALI 352
Query: 481 SAIMAIVLLP 490
S ++A++ +P
Sbjct: 353 SGLIAVLAIP 382
>BM108083
Length = 611
Score = 111 bits (277), Expect = 9e-25
Identities = 62/105 (59%), Positives = 71/105 (67%), Gaps = 2/105 (1%)
Frame = +3
Query: 213 TDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGDLFGAF 272
T +C FCANLKSCFFFSILLLL L+ AL+YV+D + P D P SCF LFGA
Sbjct: 3 TSSC-RFCANLKSCFFFSILLLLFLATVALLYVKDKQVEAIPLDDATQP-SCFFQLFGAL 176
Query: 273 KELKKPMWILMLVTAVNW--VAWFPFFLFDTDWMGREVYGGVVGE 315
KELK+PMW+LMLVTAVNW V + F T W+ EVYGG GE
Sbjct: 177 KELKRPMWMLMLVTAVNWVGVGFLTSFSTLTGWV-LEVYGGTAGE 308
Score = 69.3 bits (168), Expect = 4e-12
Identities = 44/75 (58%), Positives = 49/75 (64%), Gaps = 4/75 (5%)
Frame = +2
Query: 293 WFPFFLFDTDWMGR-EVYGGVVGEKAYDDGVRA-GSLGLMINAV-VLAFMSLAVEPLG-R 348
WFP+FLFDTDWMG + G Y G A SLGLM+NAV V A SLAVEP+G R
Sbjct: 239 WFPYFLFDTDWMGP*GLRWNCWGXTPYAXGNCAWDSLGLMVNAVLVWASCSLAVEPVGER 418
Query: 349 ILGGVKNLWGIVNFI 363
LG +K LW IVNFI
Sbjct: 419 XLGELKKLWAIVNFI 463
>TC222062 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, partial (13%)
Length = 428
Score = 87.8 bits (216), Expect = 1e-17
Identities = 40/61 (65%), Positives = 51/61 (83%)
Frame = +1
Query: 430 AGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLL 489
+G GQGL++GVLNLAIVVPQM++S SGPWDALFGGGN+PAFV+ +V A A++A + L
Sbjct: 7 SGGGQGLAIGVLNLAIVVPQMIISLGSGPWDALFGGGNIPAFVLASVCALAGAVIATLKL 186
Query: 490 P 490
P
Sbjct: 187 P 189
>TC234069 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, partial (12%)
Length = 612
Score = 82.0 bits (201), Expect = 6e-16
Identities = 36/58 (62%), Positives = 48/58 (82%)
Frame = +3
Query: 433 GQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
GQGL++GVLNLAIV+PQM++S SGPWDALFGGGN+PAFV+ ++ A ++A + LP
Sbjct: 15 GQGLAIGVLNLAIVIPQMIISLGSGPWDALFGGGNIPAFVLASLCALAGGVIATLKLP 188
>BM095066
Length = 300
Score = 55.8 bits (133), Expect = 4e-08
Identities = 32/59 (54%), Positives = 39/59 (65%), Gaps = 4/59 (6%)
Frame = +1
Query: 350 LGGVKNLWGIVNFILAICMAMTVLITKTAEHDR---LISGGATVGAPSSG-VKAGAIAF 404
L GVK LWGIVNF+LA+C+AMTVL+TK +H R L+S SSG AGA+ F
Sbjct: 43 LVGVKRLWGIVNFLLAVCLAMTVLVTKMFQHSRQYTLLSYAHQEHLASSGFFLAGALXF 219
>CA801514 homologue to GP|7649151|gb|A sucrose transport protein {Euphorbia
esula}, partial (3%)
Length = 409
Score = 32.7 bits (73), Expect = 0.40
Identities = 16/21 (76%), Positives = 18/21 (85%)
Frame = +2
Query: 435 GLSLGVLNLAIVVPQMLVSTL 455
GLSLGVLNLAIVVPQ+ + L
Sbjct: 212 GLSLGVLNLAIVVPQVRIHFL 274
>TC233237
Length = 420
Score = 32.7 bits (73), Expect = 0.40
Identities = 15/17 (88%), Positives = 17/17 (99%)
Frame = +1
Query: 434 QGLSLGVLNLAIVVPQM 450
+GLSLGVLNLAIVVPQ+
Sbjct: 274 EGLSLGVLNLAIVVPQV 324
>TC232346 similar to UP|Q96469 (Q96469) Calcium dependent protein kinase ,
partial (5%)
Length = 521
Score = 32.3 bits (72), Expect = 0.52
Identities = 15/16 (93%), Positives = 16/16 (99%)
Frame = +3
Query: 434 QGLSLGVLNLAIVVPQ 449
+GLSLGVLNLAIVVPQ
Sbjct: 135 EGLSLGVLNLAIVVPQ 182
>AW595987
Length = 410
Score = 30.4 bits (67), Expect = 2.0
Identities = 12/34 (35%), Positives = 21/34 (61%)
Frame = -1
Query: 387 GATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPF 420
G +G+ + G+ G + F+G + IPL +N+ V F
Sbjct: 164 GVAIGSTAGGI-CGVVPFYGCIQIPLTLNYQVCF 66
>TC223632
Length = 432
Score = 30.4 bits (67), Expect = 2.0
Identities = 17/36 (47%), Positives = 19/36 (52%)
Frame = +2
Query: 415 NFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQM 450
NF P + LSLGVLNLAIVVPQ+
Sbjct: 167 NFK*PIGFYTFTDLNFFLASSLSLGVLNLAIVVPQV 274
>BE609235 weakly similar to SP|P08926|RUBA RuBisCO subunit binding-protein
alpha subunit chloroplast precursor, partial (25%)
Length = 450
Score = 29.6 bits (65), Expect = 3.4
Identities = 12/37 (32%), Positives = 22/37 (59%)
Frame = -2
Query: 468 LPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLP 504
+ AF++G+ M V+ ++ + P P +DVA +S P
Sbjct: 245 ISAFILGSAMIVVALLVIVTFRPVPSWSDVAPSSVRP 135
>TC217263 similar to UP|Q949X7 (Q949X7) Diaminopimelate decarboxylase,
partial (27%)
Length = 665
Score = 29.6 bits (65), Expect = 3.4
Identities = 13/43 (30%), Positives = 21/43 (48%)
Frame = +1
Query: 31 RKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWL 73
R + V S+ G+ GW L+L+ V +G+ W + WL
Sbjct: 490 RTWVVVLSLLLGMILGWLFVLALIPRGVSLMGMGKSWRIWSWL 618
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,322,113
Number of Sequences: 63676
Number of extensions: 345413
Number of successful extensions: 2644
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 2601
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2629
length of query: 510
length of database: 12,639,632
effective HSP length: 101
effective length of query: 409
effective length of database: 6,208,356
effective search space: 2539217604
effective search space used: 2539217604
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0134.5


