

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.21
(515 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
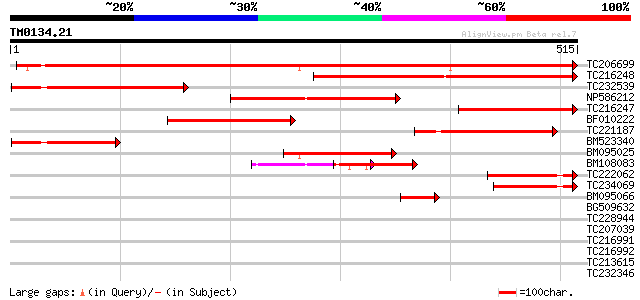
Score E
Sequences producing significant alignments: (bits) Value
TC206699 UP|Q7XA53 (Q7XA53) Sucrose transporter, complete 687 0.0
TC216248 similar to UP|O04077 (O04077) Sucrose transport protein... 346 2e-95
TC232539 similar to UP|Q7XA53 (Q7XA53) Sucrose transporter, part... 239 2e-63
NP586212 hypothetical protein [Glycine max] 218 5e-57
TC216247 similar to UP|O04077 (O04077) Sucrose transport protein... 166 3e-41
BF010222 similar to GP|28172870|em sucrose transporter 4 protein... 132 3e-31
TC221187 similar to UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 pro... 129 2e-30
BM523340 128 5e-30
BM095025 122 3e-28
BM108083 92 7e-19
TC222062 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, pa... 91 1e-18
TC234069 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, pa... 86 5e-17
BM095066 53 4e-07
BG509632 33 0.31
TC228944 33 0.40
TC207039 32 0.52
TC216991 similar to UP|Q6NM15 (Q6NM15) At1g30475, partial (19%) 32 0.52
TC216992 similar to UP|Q6NM15 (Q6NM15) At1g30475, partial (48%) 32 0.52
TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase ... 28 0.55
TC232346 similar to UP|Q96469 (Q96469) Calcium dependent protein... 32 0.68
>TC206699 UP|Q7XA53 (Q7XA53) Sucrose transporter, complete
Length = 2053
Score = 687 bits (1772), Expect = 0.0
Identities = 344/520 (66%), Positives = 411/520 (78%), Gaps = 11/520 (2%)
Frame = +1
Query: 7 PPNSTTQN---NNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTP 63
P +ST QN NN++ S VE PSPL K+I VASIAAGVQFGWALQLSLLTP
Sbjct: 22 PFSSTKQNHNNNNTLTKPSLHVESPP--LEPSPLRKIIVVASIAAGVQFGWALQLSLLTP 195
Query: 64 YVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVF 123
YVQLLG+PH W+++IWLCGPISG++VQPIVGY SDRCTSRFGRRRPFI AG+ AVAIAVF
Sbjct: 196 YVQLLGIPHTWAAYIWLCGPISGMLVQPIVGYHSDRCTSRFGRRRPFIAAGSFAVAIAVF 375
Query: 124 LIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSR 183
LIG+AAD+G+S GDDLSKK RPRA+ FV+GFWILDVANNMLQGPCRA LGDL G+H +
Sbjct: 376 LIGYAADLGHSFGDDLSKKVRPRAIGIFVVGFWILDVANNMLQGPCRALLGDLCAGNHQK 555
Query: 184 IRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLV 243
R AN FSFFM VGNVLGY AG+Y L+ +FPFT+T ACDV+CANLKSCFF SI LL
Sbjct: 556 TRNANAFFSFFMAVGNVLGYAAGAYSKLYHVFPFTKTTACDVYCANLKSCFFLSIALLTT 735
Query: 244 LSGFALFYVHDPPIGSRR---EEDDKAPKNVFVELFGAFKELKKPMLMLMLVTSLNWIAW 300
L+ AL YV + P+ + + DD F +LFGAF+ELK+PM +L+LVT LNWIAW
Sbjct: 736 LATAALVYVKEVPLSPEKAVIDSDDNGGMPCFGQLFGAFRELKRPMWILLLVTCLNWIAW 915
Query: 301 FPYVLYDTDWMGLEVYGGKLG-SKAYDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGG 359
FP++L+DTDWMG EVY G +G KAYD GVRAGALGL+LNSVVLG SL VE L R +GG
Sbjct: 916 FPFLLFDTDWMGREVYEGTVGEGKAYDRGVRAGALGLMLNSVVLGATSLGVEVLARGVGG 1095
Query: 360 VKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGR----PTPGVKAGALMFFAVL 415
VKRLW IVN +LAVC+AMT+L+TK+A+H R+ + + P VKAGAL F++L
Sbjct: 1096VKRLWGIVNFLLAVCLAMTVLVTKMAQHSRQYTLLPNAHQEPLPPPAAVKAGALALFSLL 1275
Query: 416 GIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNL 475
GIPLAITYS+PFALASI+SS++GAGQGLSLGVLN+AIVIPQM+VS ++GPWD+LFGGGNL
Sbjct: 1276GIPLAITYSIPFALASIFSSTSGAGQGLSLGVLNLAIVIPQMVVSVISGPWDALFGGGNL 1455
Query: 476 PAFMVGAVMAAVSAVLAMVLLPSPKPEEMAKASISASGFH 515
PAF+VGAV AA S +L+++LLPSP P+ A+ + GFH
Sbjct: 1456PAFVVGAVAAAASGILSIILLPSPPPDLAKAATAAGGGFH 1575
Score = 28.1 bits (61), Expect = 9.9
Identities = 11/34 (32%), Positives = 20/34 (58%)
Frame = -3
Query: 5 PPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLA 38
PP P ++T N +APS+ + + +A +PL+
Sbjct: 1094 PPTPRASTSTPNDVAPSTTEFNISPNAPARTPLS 993
>TC216248 similar to UP|O04077 (O04077) Sucrose transport protein, partial
(41%)
Length = 1234
Score = 346 bits (887), Expect = 2e-95
Identities = 169/240 (70%), Positives = 205/240 (85%), Gaps = 1/240 (0%)
Frame = +1
Query: 277 GAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGSKAYDAGVRAGALGL 336
GA KELK+PM MLMLVT++NW+ WFPY L+DTDWMG EVYGG G AY GVR G+LGL
Sbjct: 1 GALKELKRPMWMLMLVTAVNWVGWFPYFLFDTDWMGREVYGGTAGEDAYAKGVRVGSLGL 180
Query: 337 VLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGAT 396
++N+VVLG MSLAVEPLG+ +GGVKRLWAIVN ILA+ MT++ITKVAEH RR++ A
Sbjct: 181 MVNAVVLGFMSLAVEPLGKMVGGVKRLWAIVNFILAIGFGMTVVITKVAEHQRRMNPAA- 357
Query: 397 IGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQ 456
+G P+ GV G+++FF VLG+PLAIT+SVPFALASIY S++GAGQGLSLGVLN+AIV+PQ
Sbjct: 358 VGHPSEGVVVGSMVFFGVLGVPLAITFSVPFALASIYCSASGAGQGLSLGVLNLAIVVPQ 537
Query: 457 MIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKPEEMAKA-SISASGFH 515
M+VSAL+GPWDSLFGGGNLPAFMVGA AA+SA++A+VLLP+PKP + AKA S+ A GFH
Sbjct: 538 MVVSALSGPWDSLFGGGNLPAFMVGAAAAALSAIMAIVLLPTPKPADEAKASSMMAGGFH 717
>TC232539 similar to UP|Q7XA53 (Q7XA53) Sucrose transporter, partial (25%)
Length = 591
Score = 239 bits (610), Expect = 2e-63
Identities = 120/161 (74%), Positives = 131/161 (80%)
Frame = +1
Query: 2 APPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLL 61
AP P PN T+ +N+ S + PAQ SPL KM AVASIAAG+QFGWALQLSLL
Sbjct: 106 APSPTKPNDPTKPSNTSTTLSLEAGPAQ----ASPLRKMFAVASIAAGIQFGWALQLSLL 273
Query: 62 TPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIA 121
TPYVQLLGVPH +SFIWLCGPISGLVVQPIVGY SD CTSRFGRRRPFI GA+AVA+A
Sbjct: 274 TPYVQLLGVPHAAASFIWLCGPISGLVVQPIVGYYSDHCTSRFGRRRPFILGGALAVAVA 453
Query: 122 VFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVAN 162
VFLIG+AADIGY+ GDD+SK TRPRAV FVIGFWILDVAN
Sbjct: 454 VFLIGYAADIGYAAGDDISKTTRPRAVGVFVIGFWILDVAN 576
>NP586212 hypothetical protein [Glycine max]
Length = 470
Score = 218 bits (555), Expect = 5e-57
Identities = 105/155 (67%), Positives = 124/155 (79%)
Frame = +2
Query: 201 LGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSR 260
LGY AGSY GLHK+FPFTETKACDVFCANLKSCFFFSI LLL L+ AL YV D + +R
Sbjct: 2 LGYAAGSYKGLHKMFPFTETKACDVFCANLKSCFFFSILLLLFLATVALLYVKDKQVEAR 181
Query: 261 REEDDKAPKNVFVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKL 320
+D P + F +LF A KELK+PM MLMLVT++NW+ WFPY L+DTDWMG EVYGG++
Sbjct: 182 ALDDATQP-SCFFQLFSALKELKRPMWMLMLVTAVNWVGWFPYFLFDTDWMGREVYGGQV 358
Query: 321 GSKAYDAGVRAGALGLVLNSVVLGLMSLAVEPLGR 355
G AY GVR G+LGL++N+VVLG MSLAVEPLG+
Sbjct: 359 GEDAYANGVRVGSLGLMVNAVVLGFMSLAVEPLGK 463
>TC216247 similar to UP|O04077 (O04077) Sucrose transport protein, partial
(17%)
Length = 930
Score = 166 bits (419), Expect = 3e-41
Identities = 80/109 (73%), Positives = 99/109 (90%), Gaps = 1/109 (0%)
Frame = +2
Query: 408 ALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWD 467
+++FF VLG+PLAIT+SVPFALASIY S++GAGQGLSLGVLN+AIV+PQM+VS L+GPWD
Sbjct: 2 SMVFFGVLGVPLAITFSVPFALASIYCSASGAGQGLSLGVLNLAIVVPQMVVSTLSGPWD 181
Query: 468 SLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSPKPEEMAKA-SISASGFH 515
+LFGGGNLPAFMVGA AA+SA++A+VLLP+PKP + AKA S+ A GFH
Sbjct: 182 ALFGGGNLPAFMVGAAAAALSAIMAIVLLPTPKPADEAKASSMMAGGFH 328
>BF010222 similar to GP|28172870|em sucrose transporter 4 protein {Lotus
japonicus}, partial (26%)
Length = 414
Score = 132 bits (333), Expect = 3e-31
Identities = 59/116 (50%), Positives = 77/116 (65%)
Frame = +1
Query: 144 RPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGY 203
RP A+ F++GFWILDVANN+ QGPCRA LGDL++ R R AN +S FM +GN+LGY
Sbjct: 13 RPAAITVFIVGFWILDVANNVTQGPCRALLGDLTSKDPRRTRVANAYYSLFMAIGNILGY 192
Query: 204 LAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGS 259
GSY G +KIF F + AC + CANLKS FF I + V + ++ H+ P+ S
Sbjct: 193 ATGSYSGWYKIFTFALSPACTISCANLKSAFFLDIAFIAVTTYISIMAAHEVPLNS 360
>TC221187 similar to UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 protein,
partial (26%)
Length = 977
Score = 129 bits (325), Expect = 2e-30
Identities = 66/130 (50%), Positives = 88/130 (66%)
Frame = +2
Query: 368 NIILAVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPF 427
NI++ VC +++T VA + G P G+ AL+ F +LG PLAITYSVP+
Sbjct: 5 NIMMTVCFLAMLVVTYVANN----MGYIGKDLPPTGIVIAALIIFTILGFPLAITYSVPY 172
Query: 428 ALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAV 487
AL S + S G GQGLS+GVLN+AIV+PQ+IVS +GPWD LFGGGN PAF V AV A +
Sbjct: 173 ALISTHIESLGLGQGLSMGVLNLAIVVPQIIVSLGSGPWDQLFGGGNSPAFAVAAVSALI 352
Query: 488 SAVLAMVLLP 497
S ++A++ +P
Sbjct: 353 SGLIAVLAIP 382
>BM523340
Length = 429
Score = 128 bits (322), Expect = 5e-30
Identities = 66/99 (66%), Positives = 74/99 (74%)
Frame = +2
Query: 2 APPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLL 61
AP PN T+ +N+ + S + P + SPL KM AVASIAAG+QFGWALQLSLL
Sbjct: 143 APSLTKPNDPTKPSNTSSTLSLEGGPGE----ASPLRKMFAVASIAAGIQFGWALQLSLL 310
Query: 62 TPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRC 100
TPYVQLLGVPH +SFIWLCGPISGLVVQPIVGY SD C
Sbjct: 311 TPYVQLLGVPHAAASFIWLCGPISGLVVQPIVGYYSDHC 427
>BM095025
Length = 335
Score = 122 bits (307), Expect = 3e-28
Identities = 64/111 (57%), Positives = 78/111 (69%), Gaps = 8/111 (7%)
Frame = +3
Query: 249 LFYVHDPPIGSRR-------EEDDKAPKNVFVELFGAFKELKKPMLMLMLVTSLNWIAWF 301
L YV + + S + E+ F +LFGAF+ELK+PM +L+LVT LNWIAWF
Sbjct: 3 LTYVKEKTVSSEKTVRSSVEEDGSHGGMPCFGQLFGAFRELKRPMWILLLVTCLNWIAWF 182
Query: 302 PYVLYDTDWMGLEVYGGKLG-SKAYDAGVRAGALGLVLNSVVLGLMSLAVE 351
P++L+DTDWMG EVY G +G KAYD GVRAGALGL+LNSVVLG SL VE
Sbjct: 183 PFLLFDTDWMGREVYEGTVGEGKAYDRGVRAGALGLMLNSVVLGATSLGVE 335
>BM108083
Length = 611
Score = 91.7 bits (226), Expect = 7e-19
Identities = 58/120 (48%), Positives = 72/120 (59%), Gaps = 7/120 (5%)
Frame = +3
Query: 220 TKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREEDDKAPKNVFVELFGAF 279
T +C FCANLKSCFFFSI LLL L+ AL YV D + + +D P + F +LFGA
Sbjct: 3 TSSCR-FCANLKSCFFFSILLLLFLATVALLYVKDKQVEAIPLDDATQP-SCFFQLFGAL 176
Query: 280 KELKKPMLMLMLVTSLNWIAWFPYVLYD--TDWMGLEVYGGKLGS-----KAYDAGVRAG 332
KELK+PM MLMLVT++NW+ + T W+ LEVYGG G K G+R G
Sbjct: 177 KELKRPMWMLMLVTAVNWVGVGFLTSFSTLTGWV-LEVYGGTAGEXRLTPKGIARGIR*G 353
Score = 62.0 bits (149), Expect = 6e-10
Identities = 41/80 (51%), Positives = 50/80 (62%), Gaps = 4/80 (5%)
Frame = +2
Query: 295 LNWIAWFPYVLYDTDWMGL-EVYGGKLGSKAYDAGVRA-GALGLVLNSV-VLGLMSLAVE 351
L W WFPY L+DTDWMG + G Y G A +LGL++N+V V SLAVE
Sbjct: 227 LGW-GWFPYFLFDTDWMGP*GLRWNCWGXTPYAXGNCAWDSLGLMVNAVLVWASCSLAVE 403
Query: 352 PLG-RYLGGVKRLWAIVNII 370
P+G R LG +K+LWAIVN I
Sbjct: 404 PVGERXLGELKKLWAIVNFI 463
>TC222062 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, partial (13%)
Length = 428
Score = 90.9 bits (224), Expect = 1e-18
Identities = 41/81 (50%), Positives = 62/81 (75%)
Frame = +1
Query: 435 SSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMV 494
+ +G GQGL++GVLN+AIV+PQMI+S +GPWD+LFGGGN+PAF++ +V A AV+A +
Sbjct: 1 ADSGGGQGLAIGVLNLAIVVPQMIISLGSGPWDALFGGGNIPAFVLASVCALAGAVIATL 180
Query: 495 LLPSPKPEEMAKASISASGFH 515
LP +++ +S ++GFH
Sbjct: 181 KLP-----DLSSSSFQSTGFH 228
>TC234069 similar to UP|Q8LPM4 (Q8LPM4) Sucrose transporter 2, partial (12%)
Length = 612
Score = 85.5 bits (210), Expect = 5e-17
Identities = 39/76 (51%), Positives = 58/76 (76%)
Frame = +3
Query: 440 GQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLLPSP 499
GQGL++GVLN+AIVIPQMI+S +GPWD+LFGGGN+PAF++ ++ A V+A + LP
Sbjct: 15 GQGLAIGVLNLAIVIPQMIISLGSGPWDALFGGGNIPAFVLASLCALAGGVIATLKLP-- 188
Query: 500 KPEEMAKASISASGFH 515
+++ +S ++GFH
Sbjct: 189 ---DLSSSSFQSTGFH 227
>BM095066
Length = 300
Score = 52.8 bits (125), Expect = 4e-07
Identities = 22/35 (62%), Positives = 30/35 (84%)
Frame = +1
Query: 356 YLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRR 390
+L GVKRLW IVN +LAVC+AMT+L+TK+ +H R+
Sbjct: 40 FLVGVKRLWGIVNFLLAVCLAMTVLVTKMFQHSRQ 144
>BG509632
Length = 388
Score = 33.1 bits (74), Expect = 0.31
Identities = 15/37 (40%), Positives = 20/37 (53%)
Frame = +1
Query: 1 MAPPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPL 37
+APPP PPNS +N++ S +QH A P L
Sbjct: 205 IAPPPLPPNSGVRNSSEALEPSVAPSVSQHNASPGEL 315
>TC228944
Length = 982
Score = 32.7 bits (73), Expect = 0.40
Identities = 16/46 (34%), Positives = 25/46 (53%)
Frame = +1
Query: 1 MAPPPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASI 46
+APPP PPNS +N++ S +QH A P L K++ + +
Sbjct: 16 VAPPPLPPNSGVRNSSEALEPSVAPSVSQHNARPGEL-KIVQITMV 150
>TC207039
Length = 1026
Score = 32.3 bits (72), Expect = 0.52
Identities = 44/169 (26%), Positives = 71/169 (41%), Gaps = 15/169 (8%)
Frame = +3
Query: 291 LVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGSKAYDAGVRAGALGLVLNSVV-------- 342
L++S WI + Y +W G+ +DAG G + VL S+V
Sbjct: 39 LISSAPWIK----IPYPLEW----------GAPTFDAGHAFGMMAAVLVSLVESTGAYKA 176
Query: 343 LGLMSLAVEPLGRYLG---GVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIG- 398
++ A P L G + + ++N + T+ + V + G IG
Sbjct: 177 ASRLASATPPPAHVLSRGIGWQGIGILLNGLFGTLTGSTVSVENVG-----LLGSNRIGS 341
Query: 399 RPTPGVKAGALMFFAVLGIPLAITYSVPFAL-ASIYSSSTG--AGQGLS 444
R V AG ++FF++LG A+ S+PF + A++Y G A GLS
Sbjct: 342 RRVIQVSAGFMIFFSMLGKFGALFASIPFPMFAAVYCVLFGIVASVGLS 488
>TC216991 similar to UP|Q6NM15 (Q6NM15) At1g30475, partial (19%)
Length = 811
Score = 32.3 bits (72), Expect = 0.52
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = -2
Query: 3 PPPPPPNSTTQNNNSIAPSS 22
PPPPPP+S+++ N + PSS
Sbjct: 585 PPPPPPSSSSRGGNPVPPSS 526
>TC216992 similar to UP|Q6NM15 (Q6NM15) At1g30475, partial (48%)
Length = 915
Score = 32.3 bits (72), Expect = 0.52
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = -3
Query: 3 PPPPPPNSTTQNNNSIAPSS 22
PPPPPP+S+++ N + PSS
Sbjct: 553 PPPPPPSSSSRGGNPVPPSS 494
>TC213615 similar to UP|Q9SUL7 (Q9SUL7) SNF1 like protein kinase
(CBL-interacting protein kinase 4), partial (45%)
Length = 674
Score = 28.5 bits (62), Expect(2) = 0.55
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Frame = +3
Query: 5 PPPPNSTTQNNNSIAPSSFQVEPAQHAA-GPSPLAKMIAVASIAA 48
PP P ST+ + + +A SS PA A+ PSP A + + ++A
Sbjct: 303 PPRPRSTSSSTSPVAASSSPSSPAAAASRSPSPAATSRSSSPLSA 437
Score = 22.3 bits (46), Expect(2) = 0.55
Identities = 7/7 (100%), Positives = 7/7 (100%)
Frame = +1
Query: 2 APPPPPP 8
APPPPPP
Sbjct: 253 APPPPPP 273
>TC232346 similar to UP|Q96469 (Q96469) Calcium dependent protein kinase ,
partial (5%)
Length = 521
Score = 32.0 bits (71), Expect = 0.68
Identities = 13/26 (50%), Positives = 20/26 (76%)
Frame = +3
Query: 434 SSSTGAGQGLSLGVLNVAIVIPQMIV 459
++ +GLSLGVLN+AIV+PQ ++
Sbjct: 114 TAKESCSEGLSLGVLNLAIVVPQPVL 191
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,029,814
Number of Sequences: 63676
Number of extensions: 404374
Number of successful extensions: 6944
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 4789
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6328
length of query: 515
length of database: 12,639,632
effective HSP length: 101
effective length of query: 414
effective length of database: 6,208,356
effective search space: 2570259384
effective search space used: 2570259384
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0134.21


