

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
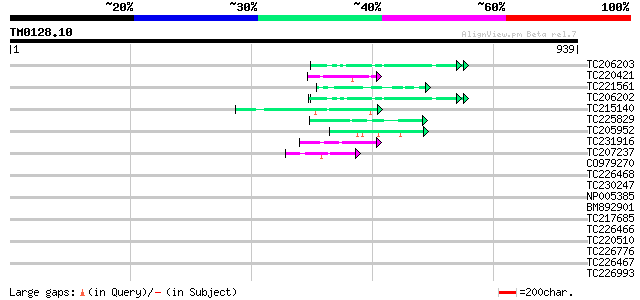
Query= TM0128.10
(939 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragme... 49 8e-06
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 47 5e-05
TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate d... 47 5e-05
TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragme... 45 1e-04
TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 45 2e-04
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 44 3e-04
TC205952 44 3e-04
TC231916 weakly similar to UP|FBRL_HUMAN (P22087) Fibrillarin (3... 44 4e-04
TC207237 weakly similar to UP|EWS_HUMAN (Q01844) RNA-binding pro... 43 6e-04
CO979270 41 0.002
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 41 0.002
TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g6... 41 0.002
NP005385 DNA-directed RNA polymerase 41 0.003
BM892901 40 0.004
TC217685 similar to GB|AAM16165.1|20334818|AY094009 AT4g26750/F1... 40 0.005
TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {... 40 0.006
TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 40 0.006
TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {A... 39 0.008
TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 39 0.008
TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 deh... 39 0.008
>TC206203 UP|Q99366 (Q99366) DNA-directed RNA polymerase (Fragment) , partial
(96%)
Length = 2604
Score = 49.3 bits (116), Expect = 8e-06
Identities = 71/266 (26%), Positives = 97/266 (35%), Gaps = 5/266 (1%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPII 557
S P TSP +SP S + P SPT +P S PTSP Y+P S
Sbjct: 1138 SSPGYSPTSPGYSPTSPGYSPTSPTYSP--------SSPGYSPTSP---AYSPTSPS--- 1275
Query: 558 HAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVS---FPTNVADSSPSNN 614
++ P+++ P S P + P+ + +S A++P S PT+ A SP++
Sbjct: 1276 YSPTSPSYSPTSPSYS----PTSPSYSPTSPSYSPTSPAYSPTSPAYSPTSPA-YSPTSP 1440
Query: 615 SESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQ 674
S S T Y T PS Y P P P + P +
Sbjct: 1441 SYS------------PTSPSYSPTSPS---YSPTSPS--YSPTSPSYSPTSPAYSPTSPG 1569
Query: 675 AHPAPDQQEPIQPEPEP-EQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGA 733
P P P P S + QS+ SP S P L S P + T SP
Sbjct: 1570 YSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSP 1749
Query: 734 SEAHNSNHPASPETNLSIVPYTHLRP 759
S + P SP + S Y+ P
Sbjct: 1750 SYS-----PTSPSYSPSSPTYSPSSP 1812
Score = 44.7 bits (104), Expect = 2e-04
Identities = 60/255 (23%), Positives = 91/255 (35%), Gaps = 5/255 (1%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPII 557
+ PA TSPS+SP S + P SP+ +P S S PTSP Y+P S
Sbjct: 1243 TSPAYSPTSPSYSPTSPSYSPTSPSYSP--------TSPSYSPTSP---SYSPTSP---A 1380
Query: 558 HAQPEPTHTEPEPR---TSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNN 614
++ P ++ P TS P + P+ + +S +++P S + + +SP+ +
Sbjct: 1381 YSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTS--PSYSPTSPAYS 1554
Query: 615 SESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGP-RPERLVDPDEPILANPLQEADPLAQ 673
S + T Y T PS P P P L +P P +
Sbjct: 1555 PTS-PGYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRL-SPSSPYSPTSP 1728
Query: 674 QAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGA 733
P P P P + SS P P S P SP
Sbjct: 1729 NYSPTSPSYSPTSPSYSPSSPTYSPSS-----PYNSGVSPDYSPSSPQFSPSTGYSPSQP 1893
Query: 734 SEAHNSNHPASPETN 748
+ +S +P+T+
Sbjct: 1894 GYSPSSTSQYTPQTS 1938
Score = 31.6 bits (70), Expect = 1.7
Identities = 29/104 (27%), Positives = 42/104 (39%), Gaps = 1/104 (0%)
Frame = +1
Query: 481 PISAMLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEE 539
P S L SS T P P TSPS+SP S + P SPT +P S
Sbjct: 1678 PSSPRLSPSSPYSPTSPNYSP----TSPSYSPTSPSYSPSSPTYSP-----------SSP 1812
Query: 540 PTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASER 583
S ++ Y+P S + P+ P ++ P+ S++
Sbjct: 1813 YNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYTPQTSDK 1944
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 46.6 bits (109), Expect = 5e-05
Identities = 42/127 (33%), Positives = 57/127 (44%), Gaps = 5/127 (3%)
Frame = +3
Query: 494 TTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSS 553
T+ P + P A TSPS SP SS PSP AP + P + PTSP +P S+
Sbjct: 291 TSSPSASPTAPPTSPSPSPPSS---PSPPSAPSPSSTAPSPPSASPPTSPSPSPPSPAST 461
Query: 554 EPI-IHAQPEPTH----TEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVAD 608
A PT T P P TS P S P+ S + TP+S + A
Sbjct: 462 ASANSPASGSPTSRTSPTSPSP-TSTSKPPAPSSSSPA*PSSKPSPSP-TPISPDPSPAT 635
Query: 609 SSPSNNS 615
S+P++++
Sbjct: 636 STPTSHT 656
Score = 42.4 bits (98), Expect = 0.001
Identities = 40/135 (29%), Positives = 57/135 (41%), Gaps = 5/135 (3%)
Frame = +3
Query: 488 HSSNPLTTIPESQPAAQTTSPSH-----SPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTS 542
H +PL T + QP +TSP H SP S+ P+ P + PS S PT+
Sbjct: 156 HHPHPLWTQNKQQPLNPSTSPPHRTPAPSPPSTM---PPSATPPSPSATSSPSAS--PTA 320
Query: 543 PITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSF 602
P T S P + P P+ T P P ++ S P + P T S+ + S
Sbjct: 321 PPTSPSPSPPSSPSPPSAPSPSSTAPSPPSA--SPPTSPSPSPPSPASTASANSPASGSP 494
Query: 603 PTNVADSSPSNNSES 617
+ + +SPS S S
Sbjct: 495 TSRTSPTSPSPTSTS 539
Score = 33.1 bits (74), Expect = 0.59
Identities = 36/125 (28%), Positives = 42/125 (32%), Gaps = 9/125 (7%)
Frame = +3
Query: 638 TCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQ----AHPAPDQQEPIQPEPEPEQ 693
T PSP P P P + P + P A A P P P P P
Sbjct: 270 TPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSPPS 449
Query: 694 SVSNQS--SVRSPHPLVETSEPH---LGTSKPNAQTFNIGSPQGASEAHNSNHPASPETN 748
S S S S P TS TSKP A P +S A S+ P+ T
Sbjct: 450 PASTASANSPASGSPTSRTSPTSPSPTSTSKPPA-------PSSSSPA*PSSKPSPSPTP 608
Query: 749 LSIVP 753
+S P
Sbjct: 609 ISPDP 623
>TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate dehydrogenase,
complete
Length = 2001
Score = 46.6 bits (109), Expect = 5e-05
Identities = 48/192 (25%), Positives = 74/192 (38%), Gaps = 2/192 (1%)
Frame = -1
Query: 508 PSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLS-SEPIIHAQPEPTHT 566
P H+P Q P E W ++ P P T Q P S S+P +Q P
Sbjct: 1182 PDHAP-----QHKPPE*YKW---------TQTPFQPSTHQPQPESDSQPGT*SQHSPQSH 1045
Query: 567 EPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSESIRKFMEVRK 626
+ + + S H+ P +++L+ +P ++PT + PS N+ +
Sbjct: 1044 QAQGKPSSHTSPSLTDKLS------------SPENYPTLQHSTQPSPNAPT--------- 928
Query: 627 EKVSTLEEYYLTCPSPRRYP-GPRPERLVDPDEPILANPLQEADPLAQQAHPAPDQQEPI 685
Y+L PR P PRP P P+ NP P P+P Q+P
Sbjct: 927 ------SPYHLPSSPPRPPP*TPRPPP-PSPAPPVSGNPSPSPPP------PSPPNQQPH 787
Query: 686 QPEPEPEQSVSN 697
P P+P+ S+
Sbjct: 786 HPSPKPQHHFSH 751
Score = 32.3 bits (72), Expect = 1.0
Identities = 34/130 (26%), Positives = 44/130 (33%), Gaps = 2/130 (1%)
Frame = -1
Query: 593 SSTAFTPVSF-PTNVADSSPSNNSE-SIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRP 650
+ T F P + P +DS P S+ S + K T SP YP +
Sbjct: 1137 TQTPFQPSTHQPQPESDSQPGT*SQHSPQSHQAQGKPSSHTSPSLTDKLSSPENYPTLQH 958
Query: 651 ERLVDPDEPILANPLQEADPLAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVET 710
P+ P L + P P P P P P P VS S P P
Sbjct: 957 STQPSPNAPTSPYHLPSSPP-----RPPP*TPRP--PPPSPAPPVSGNPSPSPPPPSPPN 799
Query: 711 SEPHLGTSKP 720
+PH + KP
Sbjct: 798 QQPHHPSPKP 769
>TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragment) ,
complete
Length = 1826
Score = 45.4 bits (106), Expect = 1e-04
Identities = 66/262 (25%), Positives = 91/262 (34%), Gaps = 1/262 (0%)
Frame = +3
Query: 499 SQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPII 557
S P TSP +SP S + P SP +P + + S PTSP Y+P S
Sbjct: 660 SSPGYSPTSPGYSPTSPGYSPTSPGYSPT-SPTYSPSSPGYSPTSP---AYSPTSPS--- 818
Query: 558 HAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSES 617
++ P+++ P S P + P+ + +S A++P S + S S S S
Sbjct: 819 YSPTSPSYSPTSPSYS----PTSPSYSPTSPSYSPTSPAYSPTSPAYSPTSPSYSPTSPS 986
Query: 618 IRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQAHP 677
T Y T PS Y P P P + P + P
Sbjct: 987 YS----------PTSPSYSPTSPS---YSPTSPS--YSPTSPAYSPTSPGYSPTSPSYSP 1121
Query: 678 APDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGASEAH 737
P P P QS+ SP S P L S P + T SP S +
Sbjct: 1122TSPSYSPTSPSYNP------QSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYS- 1280
Query: 738 NSNHPASPETNLSIVPYTHLRP 759
P SP + S Y+ P
Sbjct: 1281----PTSPSYSPSSPTYSPSSP 1334
Score = 44.3 bits (103), Expect = 3e-04
Identities = 60/259 (23%), Positives = 91/259 (34%), Gaps = 5/259 (1%)
Frame = +3
Query: 495 TIPESQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSS 553
T S P TSP++SP S + P SP+ +P S S PTSP Y+P S
Sbjct: 753 TYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSP--------TSPSYSPTSP---SYSPTSP 899
Query: 554 EPIIHAQPEPTHTEPEPR---TSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSS 610
++ P ++ P TS P + P+ + +S +++P S + + +S
Sbjct: 900 S---YSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTS--PSYSPTS 1064
Query: 611 PSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGP-RPERLVDPDEPILANPLQEAD 669
P+ + S + T Y T PS P P P L +P
Sbjct: 1065PAYSPTS-PGYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRL-SPSSPYS 1238
Query: 670 PLAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGS 729
P + P P P P + SS P P S P S
Sbjct: 1239PTSPNYSPTSPSYSPTSPSYSPSSPTYSPSS-----PYNSGVSPDYSPSSPQFSPSTGYS 1403
Query: 730 PQGASEAHNSNHPASPETN 748
P + +S +P+T+
Sbjct: 1404PSQPGYSPSSTSQYTPQTS 1460
Score = 31.6 bits (70), Expect = 1.7
Identities = 29/104 (27%), Positives = 42/104 (39%), Gaps = 1/104 (0%)
Frame = +3
Query: 481 PISAMLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEE 539
P S L SS T P P TSPS+SP S + P SPT +P S
Sbjct: 1200 PSSPRLSPSSPYSPTSPNYSP----TSPSYSPTSPSYSPSSPTYSP-----------SSP 1334
Query: 540 PTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASER 583
S ++ Y+P S + P+ P ++ P+ S++
Sbjct: 1335 YNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYTPQTSDK 1466
>TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(91%)
Length = 1885
Score = 44.7 bits (104), Expect = 2e-04
Identities = 66/265 (24%), Positives = 90/265 (33%), Gaps = 22/265 (8%)
Frame = +1
Query: 375 APEFPSPPKKKAQRKMIPEESSEESDVPLVKKKKRKPDDGDDSDNEDGPPKKKKKQVRIV 434
AP +PP + + SS S P + PP
Sbjct: 130 APNSSTPPPNSSSVPLS*SHSSSSSSSPSTTLPSP----------QTSPPTATSTPTPTS 279
Query: 435 VKPTRVEPAAEVVRRTEPPARVTRSSAHSSKSALASDD-DLNLFDALPISAMLKHSSNPL 493
P P T PP +S SS + LAS L +A +++
Sbjct: 280 SPPPPPSPPGRSRSATPPPLAAPTASPSSSPAPLASSAATAPLPSKNAATAFSGSTTSTA 459
Query: 494 TTIPE-SQPAAQT-------------TSPSHSPRSSFFQPSPTEA--PLWNLLQNQPSRS 537
TTIP + P Q T+P SS PSPT + PL L P R+
Sbjct: 460 TTIPP*NAPVRQCSGNTRFSSWKVTLTTPPCLRSSSTLSPSPTSSTSPL-RLASATPCRT 636
Query: 538 EEPTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSST-- 595
PTSP T+ + SS P P + P S S P+ + PT+ +ST
Sbjct: 637 PNPTSPPTLLASSTSSRLPNPPIPNPP*SGLHPAPSTASTPKTLSPSSTAPTNPPASTPP 816
Query: 596 ---AFTPVSFPTNVADSSPSNNSES 617
PT + +SPS +S S
Sbjct: 817 PRKPARRSPTPTTTSTASPSPDSAS 891
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 44.3 bits (103), Expect = 3e-04
Identities = 44/195 (22%), Positives = 69/195 (34%)
Frame = +2
Query: 497 PESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPI 556
P S P+ TTSP+ P + PS T +P + + P+ P T+ P SS
Sbjct: 56 PPSPPSPPTTSPACWPHTPASPPSTTTSPSPTWPKRSTAARPSPSWPSTMPPCPPSSTST 235
Query: 557 IHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSE 616
+ P T + P +S S P++S R P + P S P + P+ ++
Sbjct: 236 SPSPPSKT-SSPSTSSSTTSAPKSSTRSTTAPPSSP------PCSRPPAPPPAPPATSTS 394
Query: 617 SIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQAH 676
+ P R P P+ P P +NP ++ + +
Sbjct: 395 PTSR---------------------PARSASP-PKTTTAPSTPSTSNPSRKCPTTSPSSK 508
Query: 677 PAPDQQEPIQPEPEP 691
AP P P P
Sbjct: 509 SAPPLAPPTPRHPPP 553
Score = 33.1 bits (74), Expect = 0.59
Identities = 26/99 (26%), Positives = 35/99 (35%), Gaps = 14/99 (14%)
Frame = +2
Query: 489 SSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQY 548
+S+ T+ P+S + T PS P S P P ++P+RS P T
Sbjct: 272 TSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATSTSPTSRPARSASPPKTTTAPS 451
Query: 549 NPLSSEPIIH--------------AQPEPTHTEPEPRTS 573
P +S P A P P H P P S
Sbjct: 452 TPSTSNPSRKCPTTSPSSKSAPPLAPPTPRHPPPPPPPS 568
Score = 30.4 bits (67), Expect = 3.8
Identities = 39/176 (22%), Positives = 54/176 (30%), Gaps = 8/176 (4%)
Frame = +2
Query: 370 SRLPPAPEF-----PSPPKKKAQRKMIPEESSEESDVPLVKKKKRKPDDGDDSDNEDGPP 424
S +PP P PSPP K + P SS + P K + PP
Sbjct: 197 STMPPCPPSSTSTSPSPPSKTSS----PSTSSSTTSAP-------KSSTRSTTAPPSSPP 343
Query: 425 KKKKKQVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKSALASDDDLNLFDALPISA 484
+ T P + R PP T S S+
Sbjct: 344 CSRPPAPPPAPPATSTSPTSRPARSASPPKTTTAPSTPST-------------------- 463
Query: 485 MLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSP---TEAPLWNLLQNQPSRS 537
SNP P + P++++ P P P P T +PL +PSR+
Sbjct: 464 -----SNPSRKCPTTSPSSKSAPPLAPPTPRHPPPPPPPST*SPLCPNKAARPSRT 616
>TC205952
Length = 1414
Score = 43.9 bits (102), Expect = 3e-04
Identities = 45/193 (23%), Positives = 72/193 (36%), Gaps = 29/193 (15%)
Frame = +1
Query: 530 LQNQPSRSEEPTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSD-----HSVPRASE-- 582
+++ EE P+ + N + S+ P P T D + P AS
Sbjct: 427 IEDAKPSDEEQEPPVPVDNNVVVSDSEPAPPPPPPSAHNNFETGDLLGLNDTAPDASSIE 606
Query: 583 -----RLALKPTDTDSSTAFTPVSFPTNVAD---------SSPSNNSESIRKFMEVRKEK 628
LA+ PT+T +++AF + T D S+PS + + +
Sbjct: 607 ERNALALAIVPTETGTTSAFNTTAAQTKDFDPTGWELALVSTPSTDISAANERQLAGGLD 786
Query: 629 VSTLEEYYLTCPSPRRYP-----GPRPERLVDP---DEPILANPLQEADPLAQQAHPAPD 680
TL Y + P P P + DP I P + + QQA+P
Sbjct: 787 SLTLNSLYDEAAYRSQQPVYGAPAPNPFEMQDPFALSSSIPPPPAVQLAAMQQQANPFGP 966
Query: 681 QQEPIQPEPEPEQ 693
Q+P QP+P+P+Q
Sbjct: 967 YQQPFQPQPQPQQ 1005
>TC231916 weakly similar to UP|FBRL_HUMAN (P22087) Fibrillarin (34 kDa
nucleolar scleroderma antigen), partial (9%)
Length = 859
Score = 43.5 bits (101), Expect = 4e-04
Identities = 44/140 (31%), Positives = 57/140 (40%), Gaps = 5/140 (3%)
Frame = +1
Query: 481 PISAMLKHSSNP---LTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRS 537
P SA S+P +T PES P + SP P SS P+ P N PS S
Sbjct: 19 PTSASSSSPSSPSSLASTSPESTPLNRRPSPGSKPSSS-----PSTPPRPNPETANPSWS 183
Query: 538 EEPTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTA- 596
+ PTSP P + P + P+P PR D P + A P S+A
Sbjct: 184 QSPTSPTP----PSTMRPSANPNPKP*P*PQHPRL-DSFRPL*NPSSASNPPSAPPSSAL 348
Query: 597 -FTPVSFPTNVADSSPSNNS 615
+TP S + PS+NS
Sbjct: 349 RYTPSSPRSPTCPPGPSSNS 408
>TC207237 weakly similar to UP|EWS_HUMAN (Q01844) RNA-binding protein EWS
(EWS oncogene) (Ewing sarcoma breakpoint region 1
protein), partial (5%)
Length = 812
Score = 43.1 bits (100), Expect = 6e-04
Identities = 43/133 (32%), Positives = 57/133 (42%), Gaps = 8/133 (6%)
Frame = +3
Query: 457 TRSSAHSSKSALASDDDLNLFDALPISAMLKHSSNPLTTIPESQPAAQTTSPSHSPRS-- 514
TR S+ +S+S+ + N + P S P TT+P ++ TS S SP S
Sbjct: 309 TRRSSTTSRSSASRSSATNSPSSPP-------PSWPTTTLPTPTSSSSATSSSRSPSSPS 467
Query: 515 --SFFQPSPTEA---PLWNLLQNQPSRSEEPTSPITIQYNPLSSEPIIHAQPEPTHTEP- 568
S PSPT A P W PS TSP + P SS P + + P+ P
Sbjct: 468 PSSASTPSPTSASLGPTW--ASRTPSPWTRATSPASPTPWPSSSSPRLTSPWAPSTAPPS 641
Query: 569 EPRTSDHSVPRAS 581
P TS S P +S
Sbjct: 642 SPATSSSS*PSSS 680
>CO979270
Length = 837
Score = 41.2 bits (95), Expect = 0.002
Identities = 39/142 (27%), Positives = 60/142 (41%), Gaps = 1/142 (0%)
Frame = +3
Query: 441 EPAAEVVRR-TEPPARVTRSSAHSSKSALASDDDLNLFDALPISAMLKHSSNPLTTIPES 499
E A V ++ T P T+ S + S L FD+ SA L S L ++P
Sbjct: 258 EQAKRVTKQGTSYPTPATKGSTNEQHSQLLFRPCGKFFDSPGASAALTMPSQKLLSLPHL 437
Query: 500 QPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPIIHA 559
++P H+P PSP+ ++L+ +P P+ P + Q P +S P A
Sbjct: 438 SHLTF*SNPEHTP----IPPSPSSPSRCSILRRKP-----PSPPQSEQPEPHNSAP-QQA 587
Query: 560 QPEPTHTEPEPRTSDHSVPRAS 581
+ H + EP DH P A+
Sbjct: 588 SEQNLHPQ-EPHQLDHFQPNAA 650
Score = 36.2 bits (82), Expect = 0.069
Identities = 59/213 (27%), Positives = 85/213 (39%), Gaps = 21/213 (9%)
Frame = +3
Query: 561 PEPTHTEPEPRTS---DHSVPRAS---ERLALKPTDTDSS-------TAFTPVSFPTNVA 607
P+ H + PR S +H P+AS +L P + +S S+PT
Sbjct: 132 PKSYHGQDSPRVSWTREHPFPQASAYVHKLQQPPFASCNSWNEQAKRVTKQGTSYPTPAT 311
Query: 608 DSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLV---DPDE-PILAN 663
S +N S F K S LT PS + P L +P+ PI +
Sbjct: 312 KGS-TNEQHSQLLFRPCGKFFDSPGASAALTMPSQKLLSLPHLSHLTF*SNPEHTPIPPS 488
Query: 664 PLQEAD-PLAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPH-LGTSKPN 721
P + + ++ P+P Q E +PEP S Q+S ++ HP EPH L +PN
Sbjct: 489 PSSPSRCSILRRKPPSPPQSE----QPEPHNSAPQQASEQNLHP----QEPHQLDHFQPN 644
Query: 722 AQTFNIGSPQGA--SEAHNSNHPASPETNLSIV 752
A SPQ SE + N S T L ++
Sbjct: 645 A----AASPQPLQDSEFDSLNPSNSSNTQLHLL 731
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 41.2 bits (95), Expect = 0.002
Identities = 59/255 (23%), Positives = 86/255 (33%), Gaps = 5/255 (1%)
Frame = -2
Query: 492 PLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPL 551
P TT+P + P A TSPS S + QP A SP T P
Sbjct: 956 PPTTLPPTTPTA--TSPSSSTPLAVTQPPTVVA-----------------SPPTTSTPPT 834
Query: 552 SSEPIIHAQPEPTHTEPE-PRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSS 610
+S+P + P+P +P P+ + S P+A P S P P VA
Sbjct: 833 TSQPPANVAPKPAPVKPSAPKVAPASSPKAPPPQIPIPQPPKPSPVSPPPLPPPPVASPP 654
Query: 611 PSNNSE----SIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLQ 666
P + + K + T P+P P P P P++ P
Sbjct: 653 PLPPPKIAPTPAKTPPSPAPAKATPAPAPAPTKPAPTPSPVPPPPAATPAPAPVIEVPAP 474
Query: 667 EADPLAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFN 726
P+ + PAP + + +Q+ +P ++ S P S + T
Sbjct: 473 APAPVIEVPAPAPAHHRH-RRHRHRHRHRRHQAPAPAP-TIIRKSPPAPPDSTAESDTAP 300
Query: 727 IGSPQGASEAHNSNH 741
SP A SNH
Sbjct: 299 APSPSLNLNAAPSNH 255
Score = 29.6 bits (65), Expect = 6.5
Identities = 36/159 (22%), Positives = 53/159 (32%), Gaps = 18/159 (11%)
Frame = -2
Query: 596 AFTPVSFP-TNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYP----GPRP 650
A PV P T + ++P+ S S + V + + P P P+P
Sbjct: 977 ATVPVKLPPTTLPPTTPTATSPSSSTPLAVTQPPTVVASPPTTSTPPTTSQPPANVAPKP 798
Query: 651 ERLVDPDEPILANPLQEADPLAQQAHPAPDQQEPIQPEPEPEQSVSN------------- 697
V P P +A P Q P P + P+ P P P V++
Sbjct: 797 AP-VKPSAPKVAPASSPKAPPPQIPIPQPPKPSPVSPPPLPPPPVASPPPLPPPKIAPTP 621
Query: 698 QSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGASEA 736
+ SP P T P +KP + P A+ A
Sbjct: 620 AKTPPSPAPAKATPAPAPAPTKPAPTPSPVPPPPAATPA 504
>TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g61870
{Arabidopsis thaliana;} , partial (56%)
Length = 1291
Score = 41.2 bits (95), Expect = 0.002
Identities = 38/147 (25%), Positives = 57/147 (37%)
Frame = +1
Query: 423 PPKKKKKQVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKSALASDDDLNLFDALPI 482
PP + P +PAA T R TRS+ SS SA+ L L
Sbjct: 58 PPPPSSPRTHPPP*PQSKKPAAP---STSSNRRPTRSA--SSTSAVPPPSPLTLTSTAVP 222
Query: 483 SAMLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTS 542
S + SS P TT P S P++ + P+ + + P+P+ + + PS TS
Sbjct: 223 SPLPSPSSPPPTTSPASAPSSTISKPAPTSATKNSSPTPSSSTARPICSITPSAPSPKTS 402
Query: 543 PITIQYNPLSSEPIIHAQPEPTHTEPE 569
P P ++ + P T + E
Sbjct: 403 PPLAPSKP*IPSSLLPSSPRTTKSSLE 483
Score = 30.0 bits (66), Expect = 5.0
Identities = 46/183 (25%), Positives = 68/183 (37%), Gaps = 13/183 (7%)
Frame = +1
Query: 495 TIPESQPAAQTTSPSHSPRSSFFQPSP------TEAPLWNLLQNQPSRSEE------PTS 542
T S +A T P SPR+ P P AP + +P+RS P S
Sbjct: 25 TCQSSLASATTPPPPSSPRT---HPPP*PQSKKPAAPSTSS-NRRPTRSASSTSAVPPPS 192
Query: 543 PITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSF 602
P+T+ + S P P+ + P P TS S P SST P
Sbjct: 193 PLTLTSTAVPS-------PLPSPSSPPPTTSPASAP--------------SSTISKPA-- 303
Query: 603 PTNVA-DSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPIL 661
PT+ +SSP+ +S + R + + PSP+ P P + P +
Sbjct: 304 PTSATKNSSPTPSSSTARPICSITP-----------SAPSPKTSPPLAPSKP*IPSSLLP 450
Query: 662 ANP 664
++P
Sbjct: 451 SSP 459
>NP005385 DNA-directed RNA polymerase
Length = 2934
Score = 40.8 bits (94), Expect = 0.003
Identities = 64/249 (25%), Positives = 85/249 (33%), Gaps = 2/249 (0%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPII 557
S P TSP +SP S + P SPT +P S PTSP Y+P S
Sbjct: 2221 SSPGYSPTSPGYSPTSPGYSPTSPTYSP--------SSPGYSPTSP---AYSPTS----- 2352
Query: 558 HAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSES 617
P+++ P S S +S A PT S +++P S + +SPS
Sbjct: 2353 -----PSYSPTSPAYSPTSPAYSSTSPAYSPT----SPSYSPTS--PAYSPTSPS----- 2484
Query: 618 IRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQAHP 677
Y LT PS P P + P + P
Sbjct: 2485 -----------------YSLTSPS------------YSPTSPSYSPTSPSYSPTSPAYSP 2577
Query: 678 APDQQEPIQPEPEP-EQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGASEA 736
P P P S + QS+ SP S P L + PN ++ SP S
Sbjct: 2578 TSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPTSPN---YSPTSP-SYSPT 2745
Query: 737 HNSNHPASP 745
S P+SP
Sbjct: 2746 SPSYSPSSP 2772
Score = 39.3 bits (90), Expect = 0.008
Identities = 38/124 (30%), Positives = 60/124 (47%), Gaps = 5/124 (4%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPT----SPITIQYNPLSS 553
+ P+ TSPS+SP S + P SP+ +P + + S S PT SP + YNP S+
Sbjct: 2473 TSPSYSLTSPSYSPTSPSYSPTSPSYSPT-SPAYSPTSPSYSPTSPSYSPTSPSYNPQSA 2649
Query: 554 EPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSN 613
+ P ++ PR S P + P+ + +S +++P S PT + SSP N
Sbjct: 2650 K----YSPSLAYSPSSPRLS----PTSPNYSPTSPSYSPTSPSYSP-SSPT-YSPSSPYN 2799
Query: 614 NSES 617
+ S
Sbjct: 2800 SGVS 2811
Score = 37.4 bits (85), Expect = 0.031
Identities = 52/215 (24%), Positives = 75/215 (34%), Gaps = 9/215 (4%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNP-LSSEPII 557
+ P TSP +SP S + PS + + S S PTSP +P SS
Sbjct: 2242 TSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPAYSPTSPAYSSTSPA 2421
Query: 558 HAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVS---FPTNVADSSPSNN 614
++ P+++ P S P + P+ + +S +++P S PT+ A SP++
Sbjct: 2422 YSPTSPSYSPTSPAYS----PTSPSYSLTSPSYSPTSPSYSPTSPSYSPTSPA-YSPTSP 2586
Query: 615 SESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGP-RPERLVDPDEPILANPLQEADPLAQ 673
S S T Y T PS P P P L+ P +
Sbjct: 2587 SYS------------PTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPTSPNYSPTSP 2730
Query: 674 QAHPAPDQQEPIQPEPEP----EQSVSNQSSVRSP 704
P P P P VS S SP
Sbjct: 2731 SYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPTSP 2835
Score = 33.9 bits (76), Expect = 0.34
Identities = 25/98 (25%), Positives = 43/98 (43%), Gaps = 5/98 (5%)
Frame = +1
Query: 491 NPLTTIPESQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSP----IT 545
+P S P TSP++SP S + P SP+ +P S + P+SP ++
Sbjct: 2656 SPSLAYSPSSPRLSPTSPNYSPTSPSYSPTSPSYSP--------SSPTYSPSSPYNSGVS 2811
Query: 546 IQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASER 583
Y+P S + A P+ P ++ P+ S++
Sbjct: 2812 PDYSPTSPQFSPSAGYSPSQPGYSPSSTSQDTPQTSDK 2925
>BM892901
Length = 403
Score = 40.4 bits (93), Expect = 0.004
Identities = 31/113 (27%), Positives = 51/113 (44%), Gaps = 5/113 (4%)
Frame = +1
Query: 505 TTSPSHSPRSSFFQPSPTEAPLWNLLQNQ-----PSRSEEPTSPITIQYNPLSSEPIIHA 559
T++P+ +P + P+ AP + N P + + TSP T +P+S++ ++
Sbjct: 4 TSTPTPNPNPAMASEPPSAAPAASTATNPCLPKPPPK*KSGTSPTTTTTSPISTKTMLRF 183
Query: 560 QPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPS 612
E T P S+ AS L+L P S + TP S P + SSP+
Sbjct: 184 S-ESRSTTPTSSPVSSSISTAS--LSLSPLLPFSPFSSTPFSIPPTIVGSSPT 333
>TC217685 similar to GB|AAM16165.1|20334818|AY094009 AT4g26750/F10M23_90
{Arabidopsis thaliana;} , partial (57%)
Length = 1586
Score = 40.0 bits (92), Expect = 0.005
Identities = 34/130 (26%), Positives = 55/130 (42%), Gaps = 2/130 (1%)
Frame = +3
Query: 636 YLTCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQAHPAPDQQEPIQPEPEPEQSV 695
Y + P R Y P P + D P + P +Q HP P Q P P +S
Sbjct: 825 YHSPPPSRDYHSPPPSQ--DYHSPPSSQDYHPPPP-SQDYHPPPSQDY----HPPPARSE 983
Query: 696 SNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGASEAHNSNHPASPETNLSIVP-- 753
+ S + + + HLG + P+ +T + P H ++P+ E++L VP
Sbjct: 984 GSYSELYNHQQYSPENSQHLGPNYPSHETSSYSYP------HFQSYPSFTESSLPSVPSN 1145
Query: 754 YTHLRPTSLS 763
YTH + + +S
Sbjct: 1146YTHYQGSDVS 1175
>TC226466 weakly similar to GB|AAD49970.1|5734705|F24J5 F24J5.4 {Arabidopsis
thaliana;} , partial (22%)
Length = 978
Score = 39.7 bits (91), Expect = 0.006
Identities = 48/204 (23%), Positives = 74/204 (35%), Gaps = 10/204 (4%)
Frame = +1
Query: 423 PPKKKKKQVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKSALASDDDLNLFDALPI 482
PP + + KP V P A V P T + ++ L LP
Sbjct: 175 PPTISQPPAIVAPKPAPVTPPAPKVAPASSPKAPTPQTPPPQPPKVSPVAAPTLPPPLPP 354
Query: 483 SAML--------KHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQP 534
+ K + P T P PA T +P+ +P +P+PT AP+ P
Sbjct: 355 PPVATPPPLPPPKIAPTPAKTSPAPAPAKATPAPAPAPT----KPAPTPAPI------PP 504
Query: 535 SRSEEPTSPITIQY-NPLSSEPIIHAQPEPTHTEPEPR-TSDHSVPRASERLALKPTDTD 592
+ PT + + P S + H H P P T H P A P D+
Sbjct: 505 PPTLAPTPIVEVPAPAPSSHKHRRHRHRHKRHQAPAPAPTVIHKSPPA-------PPDST 663
Query: 593 SSTAFTPVSFPTNVADSSPSNNSE 616
+ + TP P+ +++PSN+ +
Sbjct: 664 AESDTTPAPSPSLNLNAAPSNHQQ 735
Score = 37.4 bits (85), Expect = 0.031
Identities = 53/247 (21%), Positives = 86/247 (34%), Gaps = 4/247 (1%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPIIH 558
S A + P++SP ++ SP+ P + SP T P S+P
Sbjct: 49 SNAQAPASVPANSPPTTLAATSPSSGT--------PPAVTQAASPPTTSNPPTISQPPAI 204
Query: 559 AQPEPTH-TEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSES 617
P+P T P P+ + S P+A T +PV+ PT P
Sbjct: 205 VAPKPAPVTPPAPKVAPASSPKA----PTPQTPPPQPPKVSPVAAPT----LPPPLPPPP 360
Query: 618 IRKFMEVRKEKVSTLEEYYLTCPSPRR---YPGPRPERLVDPDEPILANPLQEADPLAQQ 674
+ + K++ P+P + P P P + PI P P+ +
Sbjct: 361 VATPPPLPPPKIAPTPAKTSPAPAPAKATPAPAPAPTKPAPTPAPIPPPPTLAPTPIVEV 540
Query: 675 AHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSPQGAS 734
PAP + + ++ +Q+ +P ++ S P S + T SP
Sbjct: 541 PAPAPSSHKHRRHRHRHKR---HQAPAPAP-TVIHKSPPAPPDSTAESDTTPAPSPSLNL 708
Query: 735 EAHNSNH 741
A SNH
Sbjct: 709 NAAPSNH 729
>TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (62%)
Length = 925
Score = 39.7 bits (91), Expect = 0.006
Identities = 37/122 (30%), Positives = 45/122 (36%), Gaps = 4/122 (3%)
Frame = +1
Query: 495 TIPESQPAAQTTSPSH----SPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNP 550
T P S P A TTSPS +P+SS PT + + P R PT+ T P
Sbjct: 64 TWPHSSPLAATTSPSSPPPPTPKSSANPSPPTLSSASTPFSSPPMRWVFPTASKTSTPTP 243
Query: 551 LSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSS 610
+ A P P P S P AS P +FT P+ SS
Sbjct: 244 TWTASARCAVPPPCSNPPSRT*WSSSHPIASSPTISSPGWMTWPKSFTSPDLPSTAYPSS 423
Query: 611 PS 612
PS
Sbjct: 424 PS 429
>TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {Arabidopsis
thaliana;} , partial (68%)
Length = 1307
Score = 39.3 bits (90), Expect = 0.008
Identities = 41/148 (27%), Positives = 63/148 (41%)
Frame = +2
Query: 451 EPPARVTRSSAHSSKSALASDDDLNLFDALPISAMLKHSSNPLTTIPESQPAAQTTSPSH 510
+P A T S+A S+ LA L A+P + ++ P + P S +TSPS
Sbjct: 536 KPSAPPTTSTASSASLRLAFPSAPPLVSAMPPTPT---NAPPPPSPPPSATPPSSTSPSP 706
Query: 511 SPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPIIHAQPEPTHTEPEP 570
S S+ PSP P N + + S +SP NP S+ P ++ P
Sbjct: 707 STSSTSAAPSPPSTPSSNTPSARRTSSSTSSSPKPTS-NPSSNPPSLN*TSRSITL--IP 877
Query: 571 RTSDHSVPRASERLALKPTDTDSSTAFT 598
R+S P S++ + P+ T T+ T
Sbjct: 878 RSSAT*SPPPSDKPSNNPSTTPEITSLT 961
>TC226467 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 1017
Score = 39.3 bits (90), Expect = 0.008
Identities = 58/251 (23%), Positives = 90/251 (35%), Gaps = 8/251 (3%)
Frame = +3
Query: 499 SQPAAQTTSPSHSPRSSFFQPSPTEA-PLWNLLQNQPSRSEEPT---SPITIQYNPLSSE 554
S A T+P++SP ++ SP+ PL L PT SP T P +S+
Sbjct: 39 SNAQAPATAPANSPPTTPAATSPSSGTPLAVTLP--------PTVVASPPTTSTPPTTSQ 194
Query: 555 PIIHAQPEPTH-TEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSN 613
P P+P T P P+ + S P+A +P S TP S P
Sbjct: 195 PPAIVAPKPAPVTPPAPKVAPASSPKAPPPQTPQPQPPKVSPVVTPT--------SPPPI 350
Query: 614 NSESIRKFMEVRKEKVSTLEEYYLTCPSPRR---YPGPRPERLVDPDEPILANPLQEADP 670
+ + K++ P+P + P P P + P+ P P
Sbjct: 351 PPPPVSTPPPLPPPKIAPTPAKTPPAPAPAKATPAPAPAPTKPAPTPAPVPPPPTLAPTP 530
Query: 671 LAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPNAQTFNIGSP 730
+ + PAP + + + +Q+ +P +V S P S + T SP
Sbjct: 531 VVEVPAPAPSSHKHRRHRHRHRR---HQAPAPAP-TIVRKSPPAPPDSTAESDTTPAPSP 698
Query: 731 QGASEAHNSNH 741
A +SNH
Sbjct: 699 SLNLNAASSNH 731
>TC226993 homologue to UP|GMD1_ARATH (Q9SNY3) GDP-mannose 4,6 dehydratase
(GDP-D-mannose dehydratase) (GMD) , partial (94%)
Length = 1388
Score = 39.3 bits (90), Expect = 0.008
Identities = 48/178 (26%), Positives = 65/178 (35%), Gaps = 13/178 (7%)
Frame = +2
Query: 570 PRTSDHSVPRASERLALKPTDT--DSSTAFTPVSFPTNVADSSPSNNSESIRKFMEVRKE 627
PRTS S S + PT S+T +P P + A S+PS+ + S
Sbjct: 239 PRTSTRSASTTSMWIPTTPTRPA*SSTTPISPTP-PPSAAGSTPSSPTRS---------- 385
Query: 628 KVSTLEEYYLTCPSPRRYP-------GPRPERLVDPDEPILANPLQEADPLAQQAHPAPD 680
T LT PSP RYP P P P P P A P + PAP
Sbjct: 386 ---TTSPLSLTSPSPSRYPTTPPTSSPPAPSASSRPSVPTSPPP---AAPTSATTKPAPP 547
Query: 681 Q--QEPIQPEPEPEQSVSNQSSVRSPHPLVET--SEPHLGTSKPNAQTFNIGSPQGAS 734
+ P++ P+P S + P T + L S P + + SP A+
Sbjct: 548 RCSAPPLRRSPKPPPSTPAPPTPPPNAPPTGTP*TTARLTPSSPATASSSTTSPPAAA 721
Score = 39.3 bits (90), Expect = 0.008
Identities = 40/140 (28%), Positives = 54/140 (38%), Gaps = 16/140 (11%)
Frame = +2
Query: 489 SSNPLTTIPESQPAAQT-TSPSHSPRS--SFFQPSPTEAPLWNLLQNQP-----SRSEEP 540
S+ P++ P A T +SP+ S S S PSP+ P + P SR P
Sbjct: 314 STTPISPTPPPSAAGSTPSSPTRSTTSPLSLTSPSPSRYPTTPPTSSPPAPSASSRPSVP 493
Query: 541 TSP--------ITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTD 592
TSP T P S P + P+P + P P T + P PT T
Sbjct: 494 TSPPPAAPTSATTKPAPPRCSAPPLRRSPKPPPSTPAPPTPPPNAP---------PTGTP 646
Query: 593 SSTAFTPVSFPTNVADSSPS 612
+TA S P + S+ S
Sbjct: 647 *TTARLTPSSPATASSSTTS 706
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.129 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,913,977
Number of Sequences: 63676
Number of extensions: 671579
Number of successful extensions: 6413
Number of sequences better than 10.0: 324
Number of HSP's better than 10.0 without gapping: 5066
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5973
length of query: 939
length of database: 12,639,632
effective HSP length: 106
effective length of query: 833
effective length of database: 5,889,976
effective search space: 4906350008
effective search space used: 4906350008
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0128.10


