

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0127b.3
(823 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
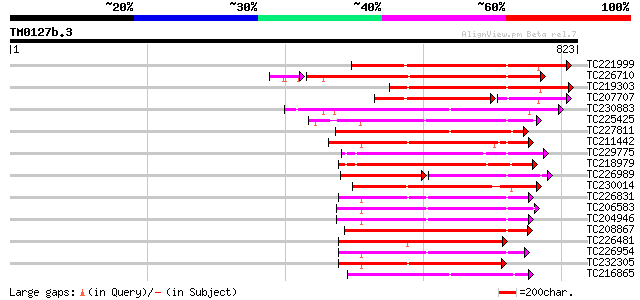
Score E
Sequences producing significant alignments: (bits) Value
TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 381 e-106
TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-l... 362 e-101
TC219303 homologue to UP|Q6W0C7 (Q6W0C7) Pto-like serine/threoni... 343 2e-94
TC207707 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 263 9e-86
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 239 3e-63
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 239 5e-63
TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , rece... 233 2e-61
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 232 6e-61
TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 226 2e-59
TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associate... 225 5e-59
TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 130 2e-57
TC230014 similar to GB|AAP21294.1|30102752|BT006486 At5g49760 {A... 220 2e-57
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 218 7e-57
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 217 1e-56
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 216 4e-56
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 215 7e-56
TC226481 similar to UP|Q41328 (Q41328) Pto kinase interactor 1, ... 214 9e-56
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 213 3e-55
TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase... 213 4e-55
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 213 4e-55
>TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (38%)
Length = 1357
Score = 381 bits (979), Expect = e-106
Identities = 192/330 (58%), Positives = 255/330 (77%), Gaps = 10/330 (3%)
Frame = +1
Query: 496 FGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERI 555
FG+VY+G +D T VAVKRG+ +S+QGLAEFRTEIEMLS+ RHRHLVSLIGYC+E+SE I
Sbjct: 1 FGRVYKGTLEDGTNVAVKRGNPRSEQGLAEFRTEIEMLSKLRHRHLVSLIGYCDERSEMI 180
Query: 556 IIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSA 615
++YEYM G L+ HL+G+ D LSWKQRLEICIGAA+GLHYLHTG++++IIHRDVK+
Sbjct: 181 LVYEYMANGPLRSHLYGT--DLPPLSWKQRLEICIGAARGLHYLHTGASQSIIHRDVKTT 354
Query: 616 NILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFG 675
NIL+D+N +AKVADFGLSKTGP +D+ +VSTAVKGSFGYLDPEY QQLTEKSDVYSFG
Sbjct: 355 NILVDDNFVAKVADFGLSKTGPALDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFG 534
Query: 676 VVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFV 735
VV+ EVLC RP ++P LPRE++N+ EW M WQ++ +++++D +L G++ SL +F
Sbjct: 535 VVLMEVLCTRPALNPVLPREQVNIAEWAMSWQKKGMLDQIMDQNL--VGKVNPASLKKFG 708
Query: 736 DTAKKCLAEHGINRPSMGDVLWHLEYALRL--------EGVDERSNH-GGDVSSQIHRSD 786
+TA+KCLAE+G++RPSMGDVLW+LEYAL+L E D +NH G +++ D
Sbjct: 709 ETAEKCLAEYGVDRPSMGDVLWNLEYALQLQETSSALMEPEDNSTNHITGIQLTRLEPFD 888
Query: 787 TGLSAME-YSMGSVGDMSNVSMSKVFAQMV 815
+S ++ + + D + + S VF+Q+V
Sbjct: 889 NTVSMIDGGNFFTDDDAEDAATSAVFSQLV 978
>TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (66%)
Length = 1737
Score = 362 bits (930), Expect(2) = e-101
Identities = 191/366 (52%), Positives = 250/366 (68%), Gaps = 18/366 (4%)
Frame = +3
Query: 431 LLCRRRSRRRNSLHIGDEKGDHG---------------ATSNYDGT-AFFTNSKIGYRLP 474
LL RRR G +G G A +N G+ A S +
Sbjct: 225 LLLLHHHRRRQGKDSGTSEGPSGWLPLSLYGNSHSAASAKTNTTGSYASSLPSNLCRHFS 404
Query: 475 LAVIQEATDNFSEDLVIGSGGFGKVYRG-VFKDETKVAVKRGSCQSQQGLAEFRTEIEML 533
A I+ AT+NF E L++G GGFGKVY+G + TKVA+KRG+ S+QG+ EF+TEIEML
Sbjct: 405 FAEIKAATNNFDEALLLGVGGFGKVYKGEIDGGTTKVAIKRGNPLSEQGVHEFQTEIEML 584
Query: 534 SQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAA 593
S+ RHRHLVSLIGYC E +E I++Y+YM G+L++HL+ + WKQRLEICIGAA
Sbjct: 585 SKLRHRHLVSLIGYCEENTEMILVYDYMAYGTLREHLYKTQKPPR--PWKQRLEICIGAA 758
Query: 594 KGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFG 653
+GLHYLHTG+ IIHRDVK+ NILLDE +AKV+DFGLSKTGP +D +VST VKGSFG
Sbjct: 759 RGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFG 938
Query: 654 YLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIE 713
YLDPEY QQLT+KSDVYSFGVV+FEVLC RP ++P+L +E+++L EW ++ ++
Sbjct: 939 YLDPEYFRRQQLTDKSDVYSFGVVLFEVLCARPALNPTLAKEQVSLAEWAAHCYQKGILD 1118
Query: 714 ELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRL-EGVDERS 772
++D +L G+I E +F +TA KC+A+ GI+RPSMGDVLW+LE+AL+L E +E
Sbjct: 1119SIIDPYL--KGKIAPECFKKFAETAMKCVADQGIDRPSMGDVLWNLEFALQLQESAEESG 1292
Query: 773 NHGGDV 778
N GD+
Sbjct: 1293NGFGDI 1310
Score = 27.3 bits (59), Expect(2) = e-101
Identities = 22/74 (29%), Positives = 36/74 (47%), Gaps = 23/74 (31%)
Frame = +2
Query: 378 EAILNGLEIMKISNSKDSL------------VFD-----SEENGKTK--VGLIVGLVAG- 417
+AILNG+EI KI+++ +L + D + +GK+K G+I G+ G
Sbjct: 20 DAILNGVEIFKINDTAGNLAGTNPIPPPVQDIIDPSMARASHHGKSKNHTGIIAGVAGGV 199
Query: 418 ---SVVGFFAIVTA 428
V+G FA +
Sbjct: 200 VLILVIGLFAFAAS 241
>TC219303 homologue to UP|Q6W0C7 (Q6W0C7) Pto-like serine/threonine kinase
(Fragment), partial (73%)
Length = 1006
Score = 343 bits (880), Expect = 2e-94
Identities = 169/276 (61%), Positives = 221/276 (79%), Gaps = 9/276 (3%)
Frame = +3
Query: 552 SERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRD 611
+E I+IYEYMEKG+LK HL+GS + LSWK+RLEICIG+A+GLHYLHTG KA+IHRD
Sbjct: 3 NEMILIYEYMEKGTLKGHLYGSGLPS--LSWKERLEICIGSARGLHYLHTGYAKAVIHRD 176
Query: 612 VKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDV 671
VKSANILLDENLMAKVADFGLSKTGP+ID+ +VSTAVKGSFGYLDPEY QQLTEKSDV
Sbjct: 177 VKSANILLDENLMAKVADFGLSKTGPEIDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDV 356
Query: 672 YSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESL 731
YSFGVV+FEVLC RPVIDP+LPRE +NL EW ++WQ++ +E+++D L +G+I+ +SL
Sbjct: 357 YSFGVVLFEVLCARPVIDPTLPREMVNLAEWALKWQKKGQLEQIIDQTL--AGKIRPDSL 530
Query: 732 HEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGV-------DERSNHGGDVSSQIH- 783
+F +TA+KCLA++G++RPSMGDVLW+LEYAL+L+ + +N G++S Q++
Sbjct: 531 RKFGETAEKCLADYGVDRPSMGDVLWNLEYALQLQEAVVQGDPEENSTNMIGELSPQVNN 710
Query: 784 -RSDTGLSAMEYSMGSVGDMSNVSMSKVFAQMVKED 818
D S +++ + D+S VSMS+VF+Q+VK +
Sbjct: 711 FNQDASASVTQFAGSGLDDLSGVSMSRVFSQLVKSE 818
>TC207707 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (34%)
Length = 1037
Score = 263 bits (671), Expect(2) = 9e-86
Identities = 125/175 (71%), Positives = 149/175 (84%)
Frame = +1
Query: 530 IEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEIC 589
+EMLS+ RH HLVSLIGYC+E+SE I++YEYM G L+ HL+G+ D LSWKQRLEIC
Sbjct: 1 VEMLSKLRHCHLVSLIGYCDERSEMILVYEYMANGPLRSHLYGT--DLPPLSWKQRLEIC 174
Query: 590 IGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVK 649
IGAA+GLHYLHTG+ ++IIHRDVK+ NILLDEN +AKVADFGLSKTGP +D+ +VSTAVK
Sbjct: 175 IGAARGLHYLHTGAAQSIIHRDVKTTNILLDENFVAKVADFGLSKTGPSLDQTHVSTAVK 354
Query: 650 GSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM 704
GSFGYLDPEY QQLTEKSDVYSFGVV+ EVLC RP ++P LPRE++N+ EW M
Sbjct: 355 GSFGYLDPEYFKRQQLTEKSDVYSFGVVLMEVLCTRPALNPVLPREQVNITEWAM 519
Score = 73.9 bits (180), Expect(2) = 9e-86
Identities = 44/118 (37%), Positives = 71/118 (59%), Gaps = 10/118 (8%)
Frame = +3
Query: 708 ERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRL-- 765
++ +++++D +L G ++ SL +F +TA+KCLAEHG++RPSMGDVLW+LEYAL+L
Sbjct: 531 KKGMLDQIMDQNLVG--KVNPASLKKFGETAEKCLAEHGVDRPSMGDVLWNLEYALQLQE 704
Query: 766 ------EGVDERSNH-GGDVSSQIHRSDTGLSAMEYSMGSV-GDMSNVSMSKVFAQMV 815
E D +NH G + + D +S ++ D +V+ S VF+Q+V
Sbjct: 705 TSSALMEPEDNSTNHITGIQLTPLDHFDNSVSMIDGGNSCTDDDTEDVATSAVFSQLV 878
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 239 bits (611), Expect = 3e-63
Identities = 156/432 (36%), Positives = 230/432 (53%), Gaps = 28/432 (6%)
Frame = +1
Query: 400 SEENGKTKVGLIVGLVAGSVVGFFAIVTAFVL--LCRRRSRRRNSLHIGDEKGDHG---- 453
S+ N G I G+V +V FF V FV + + + S G E G G
Sbjct: 1321 SKGNSSVSPGWIAGIVV--IVLFFIAVVLFVSWKCFVNKLQGKFSRVKGHENGKGGFKLD 1494
Query: 454 ---ATSNYDGTAFFTNSKI-----------GYRLPLAVIQEATDNFSEDLVIGSGGFGKV 499
++ Y G S+ G + V+Q+ T+NFSE+ ++G GGFG V
Sbjct: 1495 AVHVSNGYGGVPVELQSQSSGDRSDLHALDGPTFSIQVLQQVTNNFSEENILGRGGFGVV 1674
Query: 500 YRGVFKDETKVAVKR--GSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIII 557
Y+G D TK+AVKR +GL EF EI +LS+ RHRHLV+L+GYC ER+++
Sbjct: 1675 YKGQLHDGTKIAVKRMESVAMGNKGLKEFEAEIAVLSKVRHRHLVALLGYCINGIERLLV 1854
Query: 558 YEYMEKGSLKDHLFGSNADATC-LSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSAN 616
YEYM +G+L HLF L+WKQR+ I + A+G+ YLH+ + ++ IHRD+K +N
Sbjct: 1855 YEYMPQGTLTQHLFEWQEQGYVPLTWKQRVVIALDVARGVEYLHSLAQQSFIHRDLKPSN 2034
Query: 617 ILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGV 676
ILL +++ AKVADFGL K PD K V T + G+FGYL PEY T ++T K D+Y+FG+
Sbjct: 2035 ILLGDDMRAKVADFGLVKNAPD-GKYSVETRLAGTFGYLAPEYAATGRVTTKVDIYAFGI 2211
Query: 677 VMFEVLCGRPVIDPSLPREKMNLVEWIMR-WQERSTIEELVDHHLPGSGQIKSESLHEFV 735
V+ E++ GR +D ++P E+ +LV W R + I + +D L + ES+++
Sbjct: 2212 VLMELITGRKALDDTVPDERSHLVTWFRRVLINKENIPKAIDQTL-NPDEETMESIYKVA 2388
Query: 736 DTAKKCLAEHGINRPSMG---DVLWHLEYALRLEGVDERSN-HGGDVSSQIHRSDTGLSA 791
+ A C A RP MG +VL L + DE + GGD+ + ++ A
Sbjct: 2389 ELAGHCTAREPYQRPDMGHAVNVLVPLVEQWKPSSHDEEEDGSGGDLQMSLPQALRRWQA 2568
Query: 792 MEYSMGSVGDMS 803
E + D+S
Sbjct: 2569 NEGTSSIFNDIS 2604
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 239 bits (609), Expect = 5e-63
Identities = 137/350 (39%), Positives = 209/350 (59%), Gaps = 12/350 (3%)
Frame = +1
Query: 434 RRRSRRRNS---LHIGDEKGDHGATSNYDGTAFFTNSKIGYRLPLAVIQEATDNFSEDLV 490
R R+R NS + GD+ +G FT A ++ AT NF D V
Sbjct: 13 RNRNRYINSKFSVSSGDQPYPNGQILPTSNLRIFT---------FAELKAATRNFKADTV 165
Query: 491 IGSGGFGKVYRGVFKDE--------TKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLV 542
+G GGFGKV++G +++ T +AVK+ + +S QGL E+++E+ L + H +LV
Sbjct: 166 LGEGGFGKVFKGWLEEKATSKGGSGTVIAVKKLNSESLQGLEEWQSEVNFLGRLSHTNLV 345
Query: 543 SLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTG 602
L+GYC E+SE +++YE+M+KGSL++HLFG + L W RL+I IGAA+GL +LHT
Sbjct: 346 KLLGYCLEESELLLVYEFMQKGSLENHLFGRGSAVQPLPWDIRLKIAIGAARGLAFLHT- 522
Query: 603 SNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLIT 662
++ +I+RD K++NILLD + AK++DFGL+K GP + +V+T V G+ GY PEY+ T
Sbjct: 523 -SEKVIYRDFKASNILLDGSYNAKISDFGLAKLGPSASQSHVTTRVMGTHGYAAPEYVAT 699
Query: 663 QQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRW-QERSTIEELVDHHLP 721
L KSDVY FGVV+ E+L G +D + P + L EW+ + +R ++ ++D L
Sbjct: 700 GHLYVKSDVYGFGVVLVEILTGLRALDSNRPSGQHKLTEWVKPYLHDRRKLKGIMDSRL- 876
Query: 722 GSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGVDER 771
G+ S++ + KCLA +RPSM DVL +LE R++ +E+
Sbjct: 877 -EGKFPSKAAFRIAQLSMKCLASEPKHRPSMKDVLENLE---RIQAANEK 1014
>TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , receptor type
precursor - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (40%)
Length = 1436
Score = 233 bits (595), Expect = 2e-61
Identities = 119/285 (41%), Positives = 183/285 (63%), Gaps = 4/285 (1%)
Frame = +2
Query: 473 LPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQS--QQGLAEFRTEI 530
+ + V++ TDNFSE V+G GGFG VYRG D T++AVKR C + +G AEF++EI
Sbjct: 74 ISIQVLKNVTDNFSEKNVLGQGGFGTVYRGELHDGTRIAVKRMECGAIAGKGAAEFKSEI 253
Query: 531 EMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADAT-CLSWKQRLEIC 589
+L++ RHRHLVSL+GYC + +E++++YEYM +G+L HLF + L W +RL I
Sbjct: 254 AVLTKVRHRHLVSLLGYCLDGNEKLLVYEYMPQGTLSRHLFDWPEEGLEPLEWNRRLTIA 433
Query: 590 IGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVK 649
+ A+G+ YLH ++++ IHRD+K +NILL +++ AKVADFGL + P+ K + T +
Sbjct: 434 LDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPE-GKASIETRIA 610
Query: 650 GSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQ-E 708
G+FGYL PEY +T ++T K DV+SFGV++ E++ GR +D + P + M+LV W R
Sbjct: 611 GTFGYLAPEYAVTGRVTTKVDVFSFGVILMELITGRKALDETQPEDSMHLVTWFRRMSIN 790
Query: 709 RSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMG 753
+ + + +D + + + + S+H + A C A RP MG
Sbjct: 791 KDSFRKAIDSTIELNEETLA-SIHTVAELAGHCGAREPYQRPDMG 922
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 232 bits (591), Expect = 6e-61
Identities = 133/307 (43%), Positives = 191/307 (61%), Gaps = 10/307 (3%)
Frame = +2
Query: 464 FTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETK-------VAVKRGS 516
F SK+ Y L ++EAT++FS ++G GGFG VY+G D+ + VAVKR
Sbjct: 11 FAGSKL-YAFTLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLD 187
Query: 517 CQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNAD 576
QG E+ EI L Q RH HLV LIGYC E R+++YEYM +GSL++ LF +
Sbjct: 188 LDGLQGHREWLAEIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYSA 367
Query: 577 ATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTG 636
A + W R++I +GAAKGL +LH ++K +I+RD K++NILLD + AK++DFGL+K G
Sbjct: 368 A--MPWSTRMKIALGAAKGLAFLHE-ADKPVIYRDFKASNILLDSDFTAKLSDFGLAKDG 538
Query: 637 PDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREK 696
P+ + +V+T + G+ GY PEY++T LT KSDVYS+GVV+ E+L GR V+D S E
Sbjct: 539 PEGEDTHVTTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEG 718
Query: 697 MNLVEW---IMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMG 753
+LVEW ++R Q++ + ++D L GQ + + A KCL+ H RP+M
Sbjct: 719 KSLVEWARPLLRDQKK--VYNIIDRRL--EGQFPMKGAMKVAMLAFKCLSHHPNARPTMS 886
Query: 754 DVLWHLE 760
DV+ LE
Sbjct: 887 DVIKVLE 907
>TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (8%)
Length = 1250
Score = 226 bits (577), Expect = 2e-59
Identities = 126/301 (41%), Positives = 183/301 (59%)
Frame = +1
Query: 482 TDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHL 541
T+NF+ ++G GGFGKVY G F D+T+VAVK S + +G +F E+++L + HR+L
Sbjct: 76 TNNFTR--ILGRGGFGKVYHG-FIDDTQVAVKMLSPSAVRGYEQFLAEVKLLMRVHHRNL 246
Query: 542 VSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHT 601
SL+GYCNE++ +IYEYM G+L + + G ++ A L+W+ RL+I + AA+GL YLH
Sbjct: 247 TSLVGYCNEENNIGLIYEYMANGNLDEIVSGKSSRAKFLTWEDRLQIALDAAQGLEYLHN 426
Query: 602 GSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLI 661
G IIHRDVK ANILL+EN AK+ADFGLSK+ P Y+ST V G+ GYLDPEY I
Sbjct: 427 GCKPPIIHRDVKCANILLNENFQAKLADFGLSKSFPTDGGSYMSTVVAGTPGYLDPEYSI 606
Query: 662 TQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLP 721
+ +LTEKSDVYSFGVV+ E++ G+P I + +K ++ +W+ I+ + D
Sbjct: 607 SSRLTEKSDVYSFGVVLLEMVTGQPAI--AKTPDKTHISQWVKSMLSNGDIKNIADSRF- 777
Query: 722 GSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGVDERSNHGGDVSSQ 781
+ S+ V+ ++ RPSM ++ L+ L E + S + S
Sbjct: 778 -KEDFDTSSVWRIVEIGMASVSISPFKRPSMSYIVNELKECLTTELARKYSGRDTENSDS 954
Query: 782 I 782
I
Sbjct: 955 I 957
>TC218979 similar to UP|Q9FUU9 (Q9FUU9) Leaf senescence-associated
receptor-like protein kinase, partial (34%)
Length = 1343
Score = 225 bits (574), Expect = 5e-59
Identities = 123/289 (42%), Positives = 185/289 (63%)
Frame = +3
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFR 537
+ + T+NF+ +IG GGFG VY G + D++ VAVK S S G +F+ E+++L +
Sbjct: 174 VLKITNNFNT--IIGKGGFGTVYLG-YIDDSPVAVKVLSPSSVNGFQQFQAEVKLLVKVH 344
Query: 538 HRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLH 597
H++L SLIGYCNE + + +IYEYM G+L++HL G ++ +T LSW+ RL I + AA GL
Sbjct: 345 HKNLTSLIGYCNEGTNKALIYEYMANGNLQEHLSGKHSKSTFLSWEDRLRIAVDAALGLE 524
Query: 598 YLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDP 657
YL G IIHRDVKS NILL+E+ AK++DFGLSK P + +VST + G+ GYLDP
Sbjct: 525 YLQNGCKPPIIHRDVKSTNILLNEHFQAKLSDFGLSKAIPTDGESHVSTVLAGTPGYLDP 704
Query: 658 EYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVD 717
I+ +LT+KSDVYSFGVV+ E++ +PV++ + +EK ++ E + E+ I +VD
Sbjct: 705 HCHISSRLTQKSDVYSFGVVLLEIITNQPVMERN--QEKGHIRERVRSLIEKGDIRAIVD 878
Query: 718 HHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLE 766
L G I S + ++ A C++++ RP M ++ L+ L +E
Sbjct: 879 SRLEGDYDI--NSAWKALEIAMACVSQNPNERPIMSEIAIELKETLAIE 1019
>TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (86%)
Length = 1730
Score = 130 bits (326), Expect(2) = 2e-57
Identities = 76/181 (41%), Positives = 105/181 (57%), Gaps = 1/181 (0%)
Frame = +3
Query: 609 HRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEK 668
+RD KS+NILLDE K++DFGL+K GP DK +VST V G++GY PEY +T QLT K
Sbjct: 657 NRDCKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVK 836
Query: 669 SDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM-RWQERSTIEELVDHHLPGSGQIK 727
SDVYSFGVV E++ GR ID + P+ + NLV W + +R +L D L G+
Sbjct: 837 SDVYSFGVVFLELITGRKAIDSTQPQGEQNLVTWARPLFNDRRKFSKLADPRL--QGRFP 1010
Query: 728 SESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGVDERSNHGGDVSSQIHRSDT 787
L++ + A C+ E RP +GDV+ L Y L + D + + G + +R D
Sbjct: 1011MRGLYQALAVASMCIQESAATRPLIGDVVTALSY-LANQAYDP-NGYRGSSDDKRNRDDK 1184
Query: 788 G 788
G
Sbjct: 1185G 1187
Score = 112 bits (280), Expect(2) = 2e-57
Identities = 57/125 (45%), Positives = 78/125 (61%), Gaps = 1/125 (0%)
Frame = +2
Query: 481 ATDNFSEDLVIGSGGFGKVYRGVFKDETK-VAVKRGSCQSQQGLAEFRTEIEMLSQFRHR 539
AT NF + +G GGFG+VY+G + + VAVK+ QG EF E+ MLS H
Sbjct: 269 ATKNFRPESFVGEGGFGRVYKGRLETTAQIVAVKQLDKNGLQGNREFLVEVLMLSLLHHP 448
Query: 540 HLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYL 599
+LV+LIGYC + +R+++YE+M GSL+DHL D L W R++I +GAAKGL YL
Sbjct: 449 NLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEPLDWNTRMKIAVGAAKGLEYL 628
Query: 600 HTGSN 604
H +N
Sbjct: 629 HDKAN 643
>TC230014 similar to GB|AAP21294.1|30102752|BT006486 At5g49760 {Arabidopsis
thaliana;} , partial (29%)
Length = 1087
Score = 220 bits (561), Expect = 2e-57
Identities = 120/281 (42%), Positives = 182/281 (64%), Gaps = 6/281 (2%)
Frame = +3
Query: 498 KVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIII 557
KVY+G VA+KR + +S QG EF+TEIE+LS+ H++LV L+G+C E+ E++++
Sbjct: 3 KVYQGTLPSGELVAIKRAAKESMQGAVEFKTEIELLSRVHHKNLVGLVGFCFEKGEQMLV 182
Query: 558 YEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANI 617
YE++ G+L D L G + + W +RL++ +GAA+GL YLH ++ IIHRD+KS+NI
Sbjct: 183 YEHIPNGTLMDSLSGKSG--IWMDWIRRLKVALGAARGLAYLHELADPPIIHRDIKSSNI 356
Query: 618 LLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVV 677
LLD +L AKVADFGLSK D ++ +V+T VKG+ GYLDPEY +TQQLTEKSDVYS+GV+
Sbjct: 357 LLDHHLNAKVADFGLSKLLVDSERGHVTTQVKGTMGYLDPEYYMTQQLTEKSDVYSYGVL 536
Query: 678 MFEVLCGRPVIDPS--LPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQI----KSESL 731
M E+ R I+ + RE M +++ T ++L + H + I + + L
Sbjct: 537 MLELATARRPIEQGKYIVREVMRVMD---------TSKDLYNLHSILNPTIMKATRPKGL 689
Query: 732 HEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGVDERS 772
+FV A +C+ E+ RP+M +V+ +E + L G++ S
Sbjct: 690 EKFVMLAMRCVKEYAAERPTMAEVVKEIESMIELVGLNPNS 812
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 218 bits (556), Expect = 7e-57
Identities = 123/294 (41%), Positives = 176/294 (59%), Gaps = 11/294 (3%)
Frame = +1
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDET----------KVAVKRGSCQSQQGLAEFR 527
++ AT NF D ++G GGFG VY+G + T VAVK+ + QG E+
Sbjct: 454 LKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGLQGHKEWL 633
Query: 528 TEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLE 587
TE++ L Q H++LV LIGYC + R+++YE+M KGSL++HLF LSW R++
Sbjct: 634 TEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFRRGPQP--LSWSVRMK 807
Query: 588 ICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTA 647
+ IGAA+GL +LH ++ +I+RD K++NILLD AK++DFGL+K GP D+ +VST
Sbjct: 808 VAIGAARGLSFLHNAKSQ-VIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGDRTHVSTQ 984
Query: 648 VKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRW- 706
V G+ GY PEY+ T +LT KSDVYSFGVV+ E+L GR +D S + NLVEW +
Sbjct: 985 VMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLVEWAKPYL 1164
Query: 707 QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLE 760
++ + ++D L GQ + + A KCL RP + +VL LE
Sbjct: 1165GDKRRLFRIMDTKL--GGQYPQKGAYMAATLALKCLNREAKGRPPITEVLQTLE 1320
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 217 bits (553), Expect = 1e-56
Identities = 124/306 (40%), Positives = 181/306 (58%), Gaps = 11/306 (3%)
Frame = +1
Query: 475 LAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETK----------VAVKRGSCQSQQGLA 524
L+ ++ AT NF D V+G GGFG V++G + + +AVKR + G
Sbjct: 250 LSELKTATRNFRPDSVLGEGGFGSVFKGWIDENSLTATKPGTGIVIAVKRLNQDGIPGHR 429
Query: 525 EFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQ 584
E+ E+ L Q H HLV LIG+C E R+++YE+M +GS ++HLF + LSW
Sbjct: 430 EWLAEVNYLGQHFHSHLVRLIGFCLEDEHRLLVYEFMPRGSWENHLFRRGSYFQPLSWSL 609
Query: 585 RLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYV 644
RL++ + AAKGL +LH+ K +I+RD K++N+LLD AK++DFGL+K GP DK +V
Sbjct: 610 RLKVALDAAKGLAFLHSAEAK-VIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPTGDKSHV 786
Query: 645 STAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM 704
ST V G++GY PEYL T LT KSDVYSFGVV+ E+L G+ +D + P + NLVEW
Sbjct: 787 STRVMGTYGYAAPEYLATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHNLVEWAK 966
Query: 705 RWQ-ERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYAL 763
+ + I ++D L GQ ++ ++ A +CL+ RP+M V+ LE L
Sbjct: 967 PFMANKRKIFRVLDTRL--QGQYSTDDAYKLATLALRCLSIESKFRPNMDQVVTTLE-QL 1137
Query: 764 RLEGVD 769
+L V+
Sbjct: 1138QLSNVN 1155
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 216 bits (549), Expect = 4e-56
Identities = 123/298 (41%), Positives = 178/298 (59%), Gaps = 12/298 (4%)
Frame = +3
Query: 475 LAVIQEATDNFSEDLVIGS-GGFGKVYRGVFKDETK----------VAVKRGSCQSQQGL 523
L + AT NF +D V+G G FG V++G +++ VAVKR S S QG
Sbjct: 312 LTELTAATRNFRKDSVLGGEGDFGSVFKGWIDNQSLAAAKPGTGVVVAVKRLSLDSFQGH 491
Query: 524 AEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWK 583
+ E+ L Q H HLV LIGYC E +R+++YE+M +GSL++HLF + LSW
Sbjct: 492 KDRLAEVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLFMRGSYFQPLSWG 671
Query: 584 QRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRY 643
RL++ +GAAKGL +LH+ K +I+RD K++N+LLD N AK+AD GL+K GP +K +
Sbjct: 672 LRLKVALGAAKGLAFLHSAETK-VIYRDFKTSNVLLDSNYNAKLADLGLAKDGPTREKSH 848
Query: 644 VSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWI 703
ST V G++GY PEYL T L+ KSDV+SFGVV+ E+L GR +D + P + NLVEW
Sbjct: 849 ASTRVMGTYGYAAPEYLATGNLSAKSDVFSFGVVLLEMLSGRRAVDKNRPSGQHNLVEWA 1028
Query: 704 MRW-QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLE 760
+ + + ++D+ L GQ + + + + +CLA RP+M +V LE
Sbjct: 1029KPYLSNKRKLLRVLDNRL--EGQYELDEACKVATLSLRCLAIESKLRPTMDEVATDLE 1196
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 215 bits (547), Expect = 7e-56
Identities = 120/278 (43%), Positives = 171/278 (61%), Gaps = 4/278 (1%)
Frame = +1
Query: 486 SEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLI 545
S+ VIG GGFG VYRGV D KVA+K +QG EF+ E+E+LS+ +L++L+
Sbjct: 1 SKSNVIGHGGFGLVYRGVLNDGRKVAIKFMDQAGKQGEEEFKVEVELLSRLHSPYLLALL 180
Query: 546 GYCNEQSERIIIYEYMEKGSLKDHLFG-SNADATC--LSWKQRLEICIGAAKGLHYLHTG 602
GYC++ + ++++YE+M G L++HL+ SN+ T L W+ RL I + AAKGL YLH
Sbjct: 181 GYCSDSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLHEH 360
Query: 603 SNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLIT 662
+ +IHRD KS+NILLD+ AKV+DFGL+K GPD +VST V G+ GY+ PEY +T
Sbjct: 361 VSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYALT 540
Query: 663 QQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM-RWQERSTIEELVDHHLP 721
LT KSDVYS+GVV+ E+L GR +D P + LV W + +R + +++D L
Sbjct: 541 GHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPSL- 717
Query: 722 GSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHL 759
GQ + + + A C+ RP M DV+ L
Sbjct: 718 -EGQYSMKEVVQVAAIAAMCVQPEADYRPLMADVVQSL 828
>TC226481 similar to UP|Q41328 (Q41328) Pto kinase interactor 1, partial
(81%)
Length = 1526
Score = 214 bits (546), Expect = 9e-56
Identities = 111/250 (44%), Positives = 156/250 (62%), Gaps = 5/250 (2%)
Frame = +2
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFR 537
++E TDNF +D +IG G +G+VY GV K E A+K+ S+Q EF ++ M+S+ +
Sbjct: 389 LKEVTDNFGQDSLIGEGSYGRVYYGVLKSELAAAIKKLDA-SKQPDEEFLAQVSMVSRLK 565
Query: 538 HRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNA-----DATCLSWKQRLEICIGA 592
H + V L+GYC + S RI+ YE+ GSL D L G L+W QR++I +GA
Sbjct: 566 HENFVQLLGYCIDGSSRILAYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 745
Query: 593 AKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSF 652
A+GL YLH ++ IIHRD+KS+N+L+ ++ +AK+ADF LS PD+ R ST V G+F
Sbjct: 746 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 925
Query: 653 GYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTI 712
GY PEY +T QL KSDVYSFGVV+ E+L GR +D +LPR + +LV W +
Sbjct: 926 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 1105
Query: 713 EELVDHHLPG 722
+ VD L G
Sbjct: 1106RQCVDTRLGG 1135
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 213 bits (542), Expect = 3e-55
Identities = 120/288 (41%), Positives = 170/288 (58%), Gaps = 11/288 (3%)
Frame = +2
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETK----------VAVKRGSCQSQQGLAEFR 527
++ AT NF D V+G GGFG V++G + + VAVK+ + QG E+
Sbjct: 320 LRAATRNFRPDSVLGEGGFGSVFKGWIDEHSLAATKPGIGKIVAVKKLNQDGLQGHREWL 499
Query: 528 TEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLE 587
EI L Q +H +LV LIGYC E R+++YE+M KGS+++HLF + SW R++
Sbjct: 500 AEINYLGQLQHPNLVKLIGYCFEDEHRLLVYEFMPKGSMENHLFRRGSYFQPFSWSLRMK 679
Query: 588 ICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTA 647
I +GAAKGL +LH+ +K +I+RD K++NILLD N AK++DFGL++ GP DK +VST
Sbjct: 680 IALGAAKGLAFLHSTEHK-VIYRDFKTSNILLDTNYNAKLSDFGLARDGPTGDKSHVSTR 856
Query: 648 VKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRW- 706
V G+ GY PEYL T LT KSDVYSFGVV+ E++ GR ID + P + NLVEW +
Sbjct: 857 VMGTRGYAAPEYLATGHLTTKSDVYSFGVVLLEMISGRRAIDKNQPTGEHNLVEWAKPYL 1036
Query: 707 QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGD 754
+ + ++D L GQ A +C + RP+M +
Sbjct: 1037SNKRRVFRVMDPRL--EGQYSQNRAQAAAALAMQCFSVEPKCRPNMDE 1174
>TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase C, epsilon
type (nPKC-epsilon) , partial (5%)
Length = 908
Score = 213 bits (541), Expect = 4e-55
Identities = 115/254 (45%), Positives = 166/254 (65%), Gaps = 11/254 (4%)
Frame = +3
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDET----------KVAVKRGSCQSQQGLAEFR 527
++ AT NF D V+G GGFG+V++G T VAVK+ + S QGL E++
Sbjct: 153 LRSATRNFRPDTVLGEGGFGRVFKGWIDKNTFKPSRVGVGIPVAVKKSNPDSLQGLEEWQ 332
Query: 528 TEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLE 587
+E+++L +F H +L LIGYC E+S+ +++YEYM+KGSL+ HLF LSW RL+
Sbjct: 333 SEVQLLGKFSHPNLGKLIGYCWEESQFLLVYEYMQKGSLESHLFRRGPKP--LSWDIRLK 506
Query: 588 ICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTA 647
I IGAA+GL +LHT S K++I+RD KS+NILLD + AK++DFGL+K GP K +V+T
Sbjct: 507 IAIGAARGLAFLHT-SEKSVIYRDFKSSNILLDGDFNAKLSDFGLAKFGPVNGKSHVTTR 683
Query: 648 VKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM-RW 706
+ G++GY PEY+ T L KSDVY FGVV+ E+L GR +D + P NLVE M
Sbjct: 684 IMGTYGYAAPEYMATGHLYIKSDVYGFGVVLLEMLTGRAALDTNQPTGMQNLVECTMSSL 863
Query: 707 QERSTIEELVDHHL 720
+ ++E++D ++
Sbjct: 864 HAKKRLKEVMDPNM 905
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 213 bits (541), Expect = 4e-55
Identities = 116/270 (42%), Positives = 162/270 (59%)
Frame = +1
Query: 491 IGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNE 550
IG GGFG VY+G F D T +AVK+ S +S+QG EF EI M+S +H HLV L G C E
Sbjct: 13 IGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCCVE 192
Query: 551 QSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHR 610
+ +++YEYME SL LFG+ L W R +IC+G A+GL YLH S I+HR
Sbjct: 193 GDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIVHR 372
Query: 611 DVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSD 670
D+K+ N+LLD++L K++DFGL+K + D ++ST + G+FGY+ PEY + LT+K+D
Sbjct: 373 DIKATNVLLDQDLNPKISDFGLAKLDEE-DNTHISTRIAGTFGYMAPEYAMHGYLTDKAD 549
Query: 671 VYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSES 730
VYSFG+V E++ GR E +++EW +E+ I +LVD L G K E+
Sbjct: 550 VYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRL-GLEFNKEEA 726
Query: 731 LHEFVDTAKKCLAEHGINRPSMGDVLWHLE 760
L + A C RP+M V+ LE
Sbjct: 727 L-VMIKVALLCTNVTAALRPTMSSVVSMLE 813
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,793,300
Number of Sequences: 63676
Number of extensions: 487794
Number of successful extensions: 4119
Number of sequences better than 10.0: 934
Number of HSP's better than 10.0 without gapping: 3459
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3500
length of query: 823
length of database: 12,639,632
effective HSP length: 105
effective length of query: 718
effective length of database: 5,953,652
effective search space: 4274722136
effective search space used: 4274722136
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0127b.3


