

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
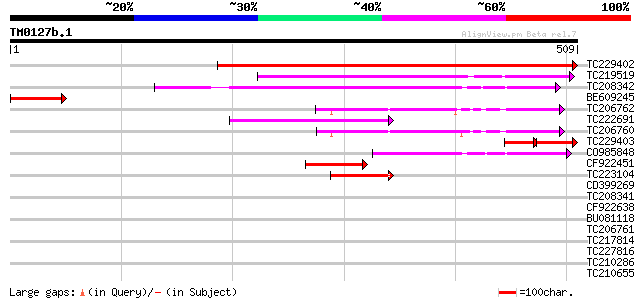
Query= TM0127b.1
(509 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229402 similar to UP|Q9SLX4 (Q9SLX4) Sigma factor, partial (63%) 577 e-165
TC219519 similar to UP|Q710C3 (Q710C3) Sigma factor (Fragment), ... 153 2e-37
TC208342 weakly similar to GB|CAB89775.1|7688083|ATH277389 sigma... 144 7e-35
BE609245 similar to GP|6682869|dbj| sigma factor {Nicotiana taba... 86 3e-17
TC206762 similar to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precurs... 79 5e-15
TC222691 weakly similar to UP|Q710C2 (Q710C2) Sigma factor (Frag... 75 9e-14
TC206760 similar to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precurs... 71 1e-12
TC229403 homologue to UP|O49935 (O49935) Sig1 protein, partial (... 70 2e-12
CO985848 62 5e-10
CF922451 53 4e-07
TC223104 similar to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precurs... 45 1e-04
CD399269 38 0.012
TC208341 weakly similar to GB|CAB89775.1|7688083|ATH277389 sigma... 36 0.047
CF922638 33 0.40
BU081118 similar to PIR|T51328|T513 transcription initiation fac... 30 2.6
TC206761 homologue to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precu... 30 3.3
TC217814 weakly similar to UP|Q6UB39 (Q6UB39) SCC3, partial (22%) 29 5.7
TC227816 29 5.7
TC210286 similar to UP|Q9FH69 (Q9FH69) Arabidopsis thaliana geno... 28 7.5
TC210655 28 9.7
>TC229402 similar to UP|Q9SLX4 (Q9SLX4) Sigma factor, partial (63%)
Length = 1079
Score = 577 bits (1487), Expect = e-165
Identities = 291/323 (90%), Positives = 309/323 (95%)
Frame = +1
Query: 187 PELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIA 246
PELLQ+R KGYVKG+VSE+LLSHAEVV LSE+IKVGLSL+EHKSRLKE+LGCEPSDDQ+A
Sbjct: 1 PELLQHRLKGYVKGIVSEELLSHAEVVNLSERIKVGLSLEEHKSRLKERLGCEPSDDQMA 180
Query: 247 ASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRG 306
SLKISR+ELRA+ IECTLARE+LAMSNVRLVMSIAQ+YDN+GAEM DLVQGGLIGLLRG
Sbjct: 181 ISLKISRTELRARMIECTLAREKLAMSNVRLVMSIAQKYDNLGAEMGDLVQGGLIGLLRG 360
Query: 307 IEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLEERGI 366
IEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLP HLHERLSLIR AKFRLEERGI
Sbjct: 361 IEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPAHLHERLSLIRNAKFRLEERGI 540
Query: 367 TPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIP 426
TPTIDRIAK LNMSQKKVRNATEAISK SLDREAFPSLNG+ G+THH YIAD+R+ENIP
Sbjct: 541 TPTIDRIAKYLNMSQKKVRNATEAISKTISLDREAFPSLNGIQGETHHSYIADDRVENIP 720
Query: 427 WHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGLSRERVRQVG 486
W+GVDEW LKDEVNKLINVTLVEREREIIRLYYGLDKE LTWEDISKRIGLSRERVRQVG
Sbjct: 721 WNGVDEWALKDEVNKLINVTLVEREREIIRLYYGLDKEGLTWEDISKRIGLSRERVRQVG 900
Query: 487 LVALEKLKHAARKREMEAMLLKH 509
LVALEKLKHAARKR+MEAMLLKH
Sbjct: 901 LVALEKLKHAARKRQMEAMLLKH 969
>TC219519 similar to UP|Q710C3 (Q710C3) Sigma factor (Fragment), partial
(56%)
Length = 1141
Score = 153 bits (387), Expect = 2e-37
Identities = 94/287 (32%), Positives = 159/287 (54%), Gaps = 2/287 (0%)
Frame = +2
Query: 223 LSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIA 282
L L++ + L E+ G +P+ Q AA + + LR + ++++ SN+RLV+SIA
Sbjct: 104 LKLEKIQEDLAERFGDQPTFAQWAAVAGVDQKTLRKRLNYGIFCKDKMIKSNIRLVISIA 283
Query: 283 QRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLR 342
+ Y G + DLVQ G GL++G EKFD +KGFK STY +WWI+Q V ++L + SRT+R
Sbjct: 284 KNYQGSGMNLQDLVQEGCRGLVKGAEKFDGTKGFKFSTYAHWWIKQAVRKSLSDQSRTIR 463
Query: 343 LPTHLHERLSLIRTAKFRL-EERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREA 401
LP H+ E ++ A+ +L E G P + +A++ +S K++ SL+++
Sbjct: 464 LPFHMVEATYRVKEARKQLYSENGRQPDDEEVAEATGLSMKRLNAVLMTPKAPRSLEQKI 643
Query: 402 FPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGL 461
+ N P + I+D E + ++ KD L +L RER+++R +G+
Sbjct: 644 GINQNLKPSEV----ISDPDAETAEEQLLKQFMKKDLEEAL--DSLNARERQVVRWRFGM 805
Query: 462 DK-ECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAMLL 507
D T ++I + +G+SRER+RQ+ A +KLK+ R ++ L+
Sbjct: 806 DDGRTKTLQEIGEMLGVSRERIRQIESSAFKKLKNKKRTNHLQQYLV 946
>TC208342 weakly similar to GB|CAB89775.1|7688083|ATH277389 sigma factor-like
protein {Arabidopsis thaliana;} , partial (41%)
Length = 1098
Score = 144 bits (364), Expect = 7e-35
Identities = 107/366 (29%), Positives = 185/366 (50%), Gaps = 2/366 (0%)
Frame = +2
Query: 131 VLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSAMQACDAMQMRSLISPELL 190
++ +Q KSSK + R+ + + ++ K D +++
Sbjct: 41 IIERRSKQRKSSKSKIIDEESYLERKSDVQRRLRLEKKLKEGYDQNDPLRL--------- 193
Query: 191 QNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLK 250
++ G S QLL+ E +L +++ +E K RL+ +L EP+ + A ++
Sbjct: 194 ------FLSGPESRQLLTREEESQLITQLQDLSRFEEVKIRLQTQLRREPTLAEWADAVG 355
Query: 251 ISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKF 310
+S L+ + ++E+L +N+R+V+ IA+ Y G + DL Q G GL++ IEKF
Sbjct: 356 LSCYTLQTQLHCANRSKEKLFHANLRMVVHIAKHYQGRGLSLQDLFQEGSTGLMKSIEKF 535
Query: 311 DSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERL-SLIRTAKFRLEERGITPT 369
G + TY YWWIR V +A+ +SRT+RLP +L+ L +I K ++E + PT
Sbjct: 536 KPEAGCRFGTYAYWWIRHAVRKAIFLHSRTIRLPENLYTLLGKVIEAKKSYIQEGNLHPT 715
Query: 370 IDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHG 429
+ +A+ + ++ +K+ N A S+ + + + T AD+ +E IP
Sbjct: 716 KEELARKVGITIEKMDNLLFASRNPISMQQTVWADQD----TTFQEITADSAIE-IPDVT 880
Query: 430 VDEWTLKDEVNKLINVTLVEREREIIRLYYGL-DKECLTWEDISKRIGLSRERVRQVGLV 488
V + ++ V+ L+N+ L +ER IIRL +G+ D E T DI K GL++ERVRQ+
Sbjct: 881 VSKQLMRMHVHNLLNI-LSPKERGIIRLRFGIEDGEEKTLSDIGKVFGLTKERVRQLESR 1057
Query: 489 ALEKLK 494
AL KL+
Sbjct: 1058ALLKLR 1075
>BE609245 similar to GP|6682869|dbj| sigma factor {Nicotiana tabacum},
partial (6%)
Length = 375
Score = 86.3 bits (212), Expect = 3e-17
Identities = 44/51 (86%), Positives = 46/51 (89%)
Frame = +3
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVA 51
MATAA IGLSGGKRLLSSSY+YSDIIEKLS SDFGSTHYQIVP KS+ VA
Sbjct: 222 MATAADIGLSGGKRLLSSSYHYSDIIEKLSYGSDFGSTHYQIVPTKSLIVA 374
>TC206762 similar to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precursor (RNA
polymerase sigma subunit SigE), partial (44%)
Length = 872
Score = 79.0 bits (193), Expect = 5e-15
Identities = 70/235 (29%), Positives = 122/235 (51%), Gaps = 11/235 (4%)
Frame = +2
Query: 275 VRLVMSIAQRYDN---MGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVS 331
+RLV+ + +Y G DL Q G+ GL+ I++F+ ++ F++STY +WIR +
Sbjct: 2 LRLVLFVINKYFTDFASGPRFQDLCQAGVKGLITAIDRFEPNRRFRLSTYSLFWIRHAII 181
Query: 332 RALVENSRTLRLPTHLHERLSLIRTAKFRLE-ERGITPTIDRIAKSLNMSQKKVRNATEA 390
R++ ++ T R+P L + I+ AK L E +PT + I + +++S ++ + +A
Sbjct: 182 RSMTLSNFT-RVPFGLESVRAEIQKAKTELTFELQRSPTEEEIIERVHISPERYHDVIKA 358
Query: 391 ISKVFSLD------REAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLIN 444
+ SL+ +E F +NG+ D DNR + + L D ++
Sbjct: 359 SKSILSLNSRHTTTQEEF--INGIVDDDG--VNGDNRKQ----PALLRLALDDVLD---- 502
Query: 445 VTLVEREREIIRLYYGLD-KECLTWEDISKRIGLSRERVRQVGLVALEKLKHAAR 498
+L +E +IR +GLD K T +I+ + +SRE VR+ + AL KLKH+AR
Sbjct: 503 -SLKPKESLVIRQRFGLDGKGDRTLGEIAGNLNISREMVRKHEVQALMKLKHSAR 664
>TC222691 weakly similar to UP|Q710C2 (Q710C2) Sigma factor (Fragment),
partial (26%)
Length = 1121
Score = 74.7 bits (182), Expect = 9e-14
Identities = 41/147 (27%), Positives = 79/147 (52%)
Frame = +1
Query: 198 VKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELR 257
V +++++ ++S+ +KV L++ ++ ++E S A + + L+
Sbjct: 175 VPNTKNKRIIVAKREAEISKGLKVLAELEKIRTAIEEDTKQVASLSNGAEASGVDEKVLQ 354
Query: 258 AKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFK 317
R+ L S LV+ +A++Y MG + DL+Q G +G+L+G +FDSS+G+K
Sbjct: 355 QLLHHGYYCRDELIQSTHSLVLYLARKYMGMGIALDDLLQAGYVGVLQGAGRFDSSRGYK 534
Query: 318 ISTYVYWWIRQGVSRALVENSRTLRLP 344
STYV +WIR+ + R + +R + +P
Sbjct: 535 FSTYVQYWIRKSILRVVARYARGIVIP 615
>TC206760 similar to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precursor (RNA
polymerase sigma subunit SigE), partial (44%)
Length = 897
Score = 70.9 bits (172), Expect = 1e-12
Identities = 67/232 (28%), Positives = 111/232 (46%), Gaps = 9/232 (3%)
Frame = +1
Query: 276 RLVMSIAQRYDN---MGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSR 332
RLV+ + +Y G DL Q G+ GL+ I++F+ + ++STY +WIR + R
Sbjct: 7 RLVLFVINKYFQDFASGPRFQDLCQAGVKGLITAIDRFEPKRRLQLSTYSLFWIRHSIVR 186
Query: 333 ALVENSRTLRLPTHLHE-RLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAI 391
++ +S T R+P L R+ + RT E +PT + + + + +S ++ +A
Sbjct: 187 SITLSSFT-RVPFGLERVRVDIQRTKLKLTFELQRSPTEEELVERIGISLERYHEVMKAS 363
Query: 392 SKVFSLDREAFPS----LNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTL 447
+ SL + +NG+ DNR + V L D ++ +L
Sbjct: 364 KPILSLHSRHITTQEEYINGITDVDG--VNGDNRRQ----LAVLRLALDDVLD-----SL 510
Query: 448 VEREREIIRLYYGLD-KECLTWEDISKRIGLSRERVRQVGLVALEKLKHAAR 498
+E +IR YGLD K T +I+ + +SRE VR+ + AL KLKH AR
Sbjct: 511 KPKESLVIRQRYGLDGKGDRTLGEIAGNLNISREIVRKHEVKALMKLKHPAR 666
>TC229403 homologue to UP|O49935 (O49935) Sig1 protein, partial (13%)
Length = 574
Score = 70.1 bits (170), Expect = 2e-12
Identities = 35/36 (97%), Positives = 36/36 (99%)
Frame = +3
Query: 474 RIGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
RIGLSRERVRQVGLVALEKLKHAARKR+MEAMLLKH
Sbjct: 252 RIGLSRERVRQVGLVALEKLKHAARKRQMEAMLLKH 359
Score = 60.5 bits (145), Expect = 2e-09
Identities = 29/30 (96%), Positives = 29/30 (96%)
Frame = +2
Query: 445 VTLVEREREIIRLYYGLDKECLTWEDISKR 474
VTLVEREREIIRLYYGLDKE LTWEDISKR
Sbjct: 2 VTLVEREREIIRLYYGLDKEGLTWEDISKR 91
>CO985848
Length = 731
Score = 62.4 bits (150), Expect = 5e-10
Identities = 52/181 (28%), Positives = 92/181 (50%), Gaps = 2/181 (1%)
Frame = -2
Query: 326 IRQGVSRALVENSRTLRLPTHLHERLSLIRTAK-FRLEERGITPTIDRIAKSLNMSQKKV 384
IRQ + +A+ +SRT+ LP + L + AK ++E + PT + +A+ + ++ +K+
Sbjct: 730 IRQAIRKAVFRHSRTIXLPEKVFILLGKVMEAKKLYIQEGNLHPTKEELARRVGVTVEKI 551
Query: 385 RNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLIN 444
+ S+ + + N T AD +E V++ ++ V +++
Sbjct: 550 DKLLFSARIPISMQQTVWADQN----TTFQEITADPTVEATDV-SVEKQLMRQHVLNVLS 386
Query: 445 VTLVEREREIIRLYYGL-DKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREME 503
+ L +ER IIRL YG D E + +I GLS+ERVRQ+ + AL KLK K+ ++
Sbjct: 385 I-LHPKERRIIRLRYGFEDGEQKSLSEIGDIFGLSKERVRQLEIRALYKLKKCLVKQGLD 209
Query: 504 A 504
A
Sbjct: 208 A 206
>CF922451
Length = 324
Score = 52.8 bits (125), Expect = 4e-07
Identities = 24/56 (42%), Positives = 36/56 (63%)
Frame = +1
Query: 266 ARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTY 321
AR L N+RLV I ++++N G + DL+ G IGL++ IE + S KG K++TY
Sbjct: 109 ARNMLIEHNLRLVAHIVKKFENTGEDAEDLISIGTIGLIKAIESYSSGKGTKLATY 276
>TC223104 similar to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precursor (RNA
polymerase sigma subunit SigE), partial (15%)
Length = 476
Score = 44.7 bits (104), Expect = 1e-04
Identities = 20/56 (35%), Positives = 36/56 (63%)
Frame = +1
Query: 289 GAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLP 344
G DL Q G+ GL+ I++F+ ++ F++STY +WIR + R++ ++ T R+P
Sbjct: 43 GPRFQDLCQAGVKGLITAIDRFEPNRRFRLSTYSLFWIRHAIIRSMTLSNFT-RVP 207
>CD399269
Length = 434
Score = 37.7 bits (86), Expect = 0.012
Identities = 24/52 (46%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Frame = -1
Query: 454 IIRLYYGL-DKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEA 504
IIRL YG D E + +I GLS+ERVRQ+ + AL KLK K+ ++A
Sbjct: 422 IIRLRYGFEDGEQKSLSEIGDIFGLSKERVRQLEIRALYKLKKCLVKQGLDA 267
>TC208341 weakly similar to GB|CAB89775.1|7688083|ATH277389 sigma factor-like
protein {Arabidopsis thaliana;} , partial (10%)
Length = 472
Score = 35.8 bits (81), Expect = 0.047
Identities = 21/52 (40%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Frame = +3
Query: 456 RLYYGL-DKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
RL +G+ D + T DI K GL++ERVRQ+ AL KLK + ++A +
Sbjct: 3 RLRFGIEDGKEKTLSDIGKVFGLTKERVRQLECRALSKLKQCLESQGLDAYI 158
>CF922638
Length = 481
Score = 32.7 bits (73), Expect = 0.40
Identities = 14/37 (37%), Positives = 21/37 (55%)
Frame = +1
Query: 294 DLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGV 330
DL+ G++GL +EKFD + K TY + IR +
Sbjct: 325 DLISLGMLGLYDALEKFDPGRDLKFDTYASFRIRGAI 435
>BU081118 similar to PIR|T51328|T513 transcription initiation factor sigma5
plastid -specific [imported] - Arabidopsis thaliana,
partial (5%)
Length = 319
Score = 30.0 bits (66), Expect = 2.6
Identities = 15/56 (26%), Positives = 32/56 (56%)
Frame = +2
Query: 229 KSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQR 284
K L+++L EP+D ++A + +S ++++ AR +L N+RLV+ + +
Sbjct: 152 KEYLQKELPTEPADGELADATNMSITQVKKAIHVGQAARNKLIKHNLRLVLFVINK 319
>TC206761 homologue to UP|Q9ZNX9 (Q9ZNX9) Sigma-like factor precursor (RNA
polymerase sigma subunit SigE), partial (6%)
Length = 317
Score = 29.6 bits (65), Expect = 3.3
Identities = 15/33 (45%), Positives = 22/33 (66%)
Frame = +2
Query: 466 LTWEDISKRIGLSRERVRQVGLVALEKLKHAAR 498
L ++ I+ + +SRE VR+ + AL KLKH AR
Sbjct: 5 LEFKQIAGNLNISREMVRKHEVKALMKLKHPAR 103
>TC217814 weakly similar to UP|Q6UB39 (Q6UB39) SCC3, partial (22%)
Length = 1311
Score = 28.9 bits (63), Expect = 5.7
Identities = 18/63 (28%), Positives = 30/63 (47%)
Frame = +1
Query: 85 AETWFQECDSNDLEVESSDLGYSVEALLLLQKSMLEKQWSLSCEREVLTEHPRQEKSSKK 144
AETWF +N + + LGY + ML+K W L ++ +++ E +K+
Sbjct: 28 AETWFLFRTTNFNKTKLEKLGYQPDT------DMLQKFWELCQQQLNISDEAEDEDVNKE 189
Query: 145 VAV 147
AV
Sbjct: 190 YAV 198
>TC227816
Length = 1221
Score = 28.9 bits (63), Expect = 5.7
Identities = 19/87 (21%), Positives = 42/87 (47%)
Frame = +2
Query: 167 GKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLD 226
GK ++ A ++R + P+ ++ + + Y LLS + L+E +K + ++
Sbjct: 557 GKPCQLVRFNSASEVRQ-VDPDYVRGQHRTYFSDGYPFLLLSQESLDALNELLKERIPIN 733
Query: 227 EHKSRLKEKLGCEPSDDQIAASLKISR 253
+ + + GC+P + + +KISR
Sbjct: 734 RFRPNILVE-GCDPYSEDLWTEIKISR 811
>TC210286 similar to UP|Q9FH69 (Q9FH69) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K16E1, partial (8%)
Length = 560
Score = 28.5 bits (62), Expect = 7.5
Identities = 19/53 (35%), Positives = 23/53 (42%)
Frame = +3
Query: 17 SSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQAFPASDRPNQ 69
SSS + ++ EKL S VP TVA SS + PA D P Q
Sbjct: 123 SSSQHQKEVAEKLGSSQ---------VPKAPYTVAGLSSQKSNPIPAEDEPMQ 254
>TC210655
Length = 723
Score = 28.1 bits (61), Expect = 9.7
Identities = 11/32 (34%), Positives = 19/32 (59%)
Frame = +3
Query: 312 SSKGFKISTYVYWWIRQGVSRALVENSRTLRL 343
+S+ F++ +V WWI +G + NS T R+
Sbjct: 321 ASR*FRVLPFVSWWIPKGKAHCASRNSFTFRV 416
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,026,568
Number of Sequences: 63676
Number of extensions: 245896
Number of successful extensions: 1173
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1163
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1167
length of query: 509
length of database: 12,639,632
effective HSP length: 101
effective length of query: 408
effective length of database: 6,208,356
effective search space: 2533009248
effective search space used: 2533009248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0127b.1


