

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0119.8
(520 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
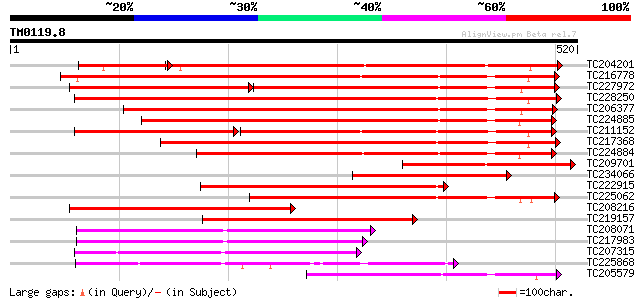
Score E
Sequences producing significant alignments: (bits) Value
TC204201 similar to UP|Q8W559 (Q8W559) Calcium/calmodulin-depend... 339 e-114
TC216778 UP|O24431 (O24431) Calmodulin-like domain protein kinas... 403 e-112
TC227972 similar to UP|Q9AR92 (Q9AR92) Protein kinase , partial... 256 e-111
TC228250 UP|Q84P29 (Q84P29) Seed calcium dependent protein kinas... 390 e-108
TC206377 similar to UP|O81390 (O81390) Calcium-dependent protein... 370 e-103
TC224885 similar to UP|Q93YI3 (Q93YI3) Calcium dependent calmodu... 361 e-100
TC211152 UP|Q84P28 (Q84P28) Seed calcium dependent protein kinas... 240 e-100
TC217368 similar to UP|Q93YF4 (Q93YF4) Calcium-dependent protein... 345 4e-95
TC224884 similar to UP|Q93YI3 (Q93YI3) Calcium dependent calmodu... 321 4e-88
TC209701 similar to UP|Q8LDS1 (Q8LDS1) Calcium-dependent protein... 275 4e-74
TC234066 similar to UP|Q8LDS1 (Q8LDS1) Calcium-dependent protein... 266 2e-71
TC222915 homologue to UP|Q84P29 (Q84P29) Seed calcium dependent ... 243 1e-64
TC225062 similar to UP|Q8W4I7 (Q8W4I7) Calcium-dependent protein... 243 2e-64
TC208216 similar to UP|O48565 (O48565) Calcium-dependent protein... 235 4e-62
TC219157 similar to UP|Q9LET1 (Q9LET1) Calcium-dependent protein... 223 1e-58
TC208071 UP|Q8GRZ5 (Q8GRZ5) Phosphoenolpyruvate carboxylase kina... 186 2e-47
TC217983 UP|Q8GSL0 (Q8GSL0) Phosphoenolpyruvate carboxylase kina... 186 2e-47
TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kina... 181 9e-46
TC225868 UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, complete 181 9e-46
TC205579 homologue to UP|Q43676 (Q43676) Calcium dependent prote... 171 7e-43
>TC204201 similar to UP|Q8W559 (Q8W559) Calcium/calmodulin-dependent protein
kinase CaMK3, partial (75%)
Length = 1757
Score = 339 bits (870), Expect(2) = e-114
Identities = 183/368 (49%), Positives = 246/368 (66%), Gaps = 4/368 (1%)
Frame = +3
Query: 144 EGGELLDRILAN--RYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSP 201
+GGELLD IL+ +Y+E DA V+ Q+L V A CHL+G+VHRD+KPENFL+ E S
Sbjct: 252 KGGELLDMILSRGGKYSEDDAKAVMVQILNVVAFCHLQGVVHRDLKPENFLYAKKDESSE 431
Query: 202 LKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPF 261
LKA DFGLSDF++P ++ +DIVGSAYYVAPEVL R G ++DVWSIGVI YILLCG RPF
Sbjct: 432 LKAIDFGLSDFVRPDERLNDIVGSAYYVAPEVLHRSYGTEADVWSIGVIAYILLCGSRPF 611
Query: 262 WDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVRE 321
W +TE GIF+ VL+ P F PWP++S AKDFVK++L KDPR R++AAQALSHPW+R
Sbjct: 612 WARTESGIFRAVLKADPSFDETPWPSLSLEAKDFVKRILNKDPRKRISAAQALSHPWIRN 791
Query: 322 GGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGS 381
++P+DI + ++ ++ S ++ ALRAL+ L +EL L++QF ++ KNGS
Sbjct: 792 YNNV-KVPLDILIFKLMKTYMRSSSLRKAALRALSKTLTADELYYLREQFALLEPSKNGS 968
Query: 382 ISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDK 441
ISLE + +AL K + +KESR+L+ L +++S +DF EF AA L VHQLE D+
Sbjct: 969 ISLENVNKALMKYATDAMKESRILDFLSSLNSLQYRRMDFEEFCAAALSVHQLEA--LDR 1142
Query: 442 WQLRSQAAFEKFDLDKDGYITPEELRMHTGLRGSI--DPLLEEADIDKDGKISLSEFRRL 499
W+ ++ A+E FD D + I EEL GL SI +L + DGK+S F +L
Sbjct: 1143WEQHARCAYELFDKDGNRAIVIEELASELGLGPSIPVHVVLHDWIRHTDGKLSFLGFVKL 1322
Query: 500 LRTASMGS 507
L S S
Sbjct: 1323LHGVSSRS 1346
Score = 90.9 bits (224), Expect(2) = e-114
Identities = 46/89 (51%), Positives = 61/89 (67%), Gaps = 3/89 (3%)
Frame = +2
Query: 64 LGKLLGHGQFGYTYVGVDKAS---GDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+G+ +G G FGYT K G +VAVK + K+KM A+EDV++EV IL+A+ GH
Sbjct: 2 VGEEVGRGHFGYTCSAKFKKGELKGQQVAVKVIPKAKMTTAIAIEDVRREVKILRALNGH 181
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELL 149
N++QFY+AF+D VYIVMELCEGG L
Sbjct: 182 SNLIQFYDAFEDQDNVYIVMELCEGGGAL 268
>TC216778 UP|O24431 (O24431) Calmodulin-like domain protein kinase isoenzyme
gamma , complete
Length = 2377
Score = 403 bits (1035), Expect = e-112
Identities = 208/474 (43%), Positives = 308/474 (64%), Gaps = 16/474 (3%)
Frame = +1
Query: 47 GMRADFGYDKDFDE---RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAA 103
G+R D K F++ Y+LGK LG GQFG TY+ + ++G + A K + K K+ +
Sbjct: 637 GVRQDTILGKQFEDVKQLYTLGKELGRGQFGVTYLCTENSTGLQYACKSISKRKLASKSD 816
Query: 104 VEDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDA 162
ED+K+E+ I++ ++G N+V+F A++D + V++VMELC GGEL DRI+A Y+EK A
Sbjct: 817 KEDIKREIQIMQHLSGQPNIVEFKGAYEDRNSVHVVMELCAGGELFDRIIAKGHYSEKAA 996
Query: 163 AVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDI 222
A + RQ++ V CH G++HRD+KPENFL S E++ LKATDFGLS FI+ GK + DI
Sbjct: 997 ASICRQIVNVVHICHFMGVMHRDLKPENFLLSSRDENALLKATDFGLSVFIEEGKVYRDI 1176
Query: 223 VGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRR 282
VGSAYYVAPEVL+R+ G + D+WS GVI YILL G PFW +TE GIF +L DF
Sbjct: 1177 VGSAYYVAPEVLRRRCGKEIDIWSAGVILYILLSGVPPFWAETEKGIFDAILEGHIDFES 1356
Query: 283 KPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFV 342
+PWP ISN AKD V+K+LI+DP+ R+T+AQ L HPW+++ G AS+ PID +VL+ ++QF
Sbjct: 1357 QPWPNISNNAKDLVRKMLIQDPKKRITSAQVLEHPWIKD-GNASDKPIDSAVLSRMKQFR 1533
Query: 343 TYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKES 402
++ K+ AL+ +A ++ EE+ LK F +D DK+G+I+ EE++ L + L KL E+
Sbjct: 1534 AMNKLKKLALKVIAENMSAEEIQGLKAMFTNMDTDKSGTITYEELKSGLHR-LGSKLTEA 1710
Query: 403 RVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYIT 462
V ++++A D + +G +D+ EF+ AT+H H+LE D + AF+ FD D G+IT
Sbjct: 1711 EVKQLMEAADVDGNGSIDYIEFITATMHRHKLERDD------QLFKAFQYFDKDNSGFIT 1872
Query: 463 PEELRMHTGLRG------------SIDPLLEEADIDKDGKISLSEFRRLLRTAS 504
+EL G +D ++ E D D DG+I+ EF ++++ +
Sbjct: 1873 RDELESAMKEYGMGDDATIKEIISEVDTIISEVDTDHDGRINYEEFSAMMKSGN 2034
>TC227972 similar to UP|Q9AR92 (Q9AR92) Protein kinase , partial (85%)
Length = 2010
Score = 256 bits (654), Expect(2) = e-111
Identities = 130/286 (45%), Positives = 187/286 (64%), Gaps = 5/286 (1%)
Frame = +3
Query: 224 GSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRK 283
GSAYYVAPEVL+R G ++D+WS GVI YILL G PFW +TE GIF +L+ DF
Sbjct: 894 GSAYYVAPEVLRRSYGKEADIWSAGVILYILLSGVPPFWAETEKGIFDAILQGHIDFESS 1073
Query: 284 PWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVT 343
PWP+ISN+AKD V+K+LIKDP+ R+TAAQ L HPW++EGG AS+ PI+ +VL+ ++QF
Sbjct: 1074 PWPSISNSAKDLVRKMLIKDPKKRITAAQVLEHPWLKEGGNASDKPINSAVLSRMKQFRA 1253
Query: 344 YSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESR 403
++ K+ AL+ +A L+EEE+ LK F ID D +G+I+ EE+R L + L KL E+
Sbjct: 1254 MNKLKKLALKVIAENLSEEEIQGLKAMFTNIDTDNSGTITYEELRAGLQR-LGSKLTETE 1430
Query: 404 VLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITP 463
V +++ A D + +G +D+ EF+ AT+H H+LE + AF+ FD D GYIT
Sbjct: 1431 VRQLMDAADVDGNGTIDYIEFITATMHRHRLERDE------HLYKAFQYFDKDGSGYITR 1592
Query: 464 EELRM-----HTGLRGSIDPLLEEADIDKDGKISLSEFRRLLRTAS 504
+EL + G +I ++ E D D DG+I+ EF ++R+ +
Sbjct: 1593 DELEIAMKEYGMGDEATIREIISEVDTDNDGRINYEEFCTMMRSGT 1730
Score = 165 bits (418), Expect(2) = e-111
Identities = 81/169 (47%), Positives = 116/169 (67%), Gaps = 1/169 (0%)
Frame = +2
Query: 56 KDFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILK 115
+D + Y+LGK LG GQFG TY+ + ++G + A K + K K+V ED+K+E+ I++
Sbjct: 386 EDVKQHYTLGKELGRGQFGVTYLCTENSTGFQYACKSISKRKLVSRDDKEDMKREIQIMQ 565
Query: 116 AVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILA-NRYTEKDAAVVVRQMLKVAA 174
++G N+V+F AF+D V++VMELC GGEL DRI+A Y+E+ AA + RQ++KV
Sbjct: 566 HLSGQSNIVEFKGAFEDKQSVHVVMELCAGGELFDRIIAKGHYSERAAASICRQIVKVVN 745
Query: 175 ECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIV 223
CH G++HRD+KPENFL S + LKATDFGLS FI+ GK + +IV
Sbjct: 746 TCHFMGVIHRDLKPENFLLSSKDDKGLLKATDFGLSVFIEEGKVYRNIV 892
>TC228250 UP|Q84P29 (Q84P29) Seed calcium dependent protein kinase a,
complete
Length = 1843
Score = 390 bits (1001), Expect = e-108
Identities = 197/452 (43%), Positives = 299/452 (65%), Gaps = 5/452 (1%)
Frame = +3
Query: 60 ERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAG 119
E Y +G+ LG GQFG T+ +ASG + A K + K K++ EDV +E+ I+ ++
Sbjct: 216 EVYEVGRKLGQGQFGTTFECTRRASGGKFACKSIPKRKLLCKEDYEDVWREIQIMHHLSE 395
Query: 120 HENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHL 178
H +VV+ ++D + V++VMELCEGGEL DRI+ Y+E+ AA +++ +++V CH
Sbjct: 396 HAHVVRIEGTYEDSTAVHLVMELCEGGELFDRIVQKGHYSERQAARLIKTIVEVVEACHS 575
Query: 179 RGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKS 238
G++HRD+KPENFLF + ED+ LKATDFGLS F KPG+ F D+VGS YYVAPEVL++
Sbjct: 576 LGVMHRDLKPENFLFDTIDEDAKLKATDFGLSVFYKPGESFCDVVGSPYYVAPEVLRKLY 755
Query: 239 GPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKK 298
GP+SDVWS GVI YILL G PFW ++E GIF+++L K DF +PWP+IS++AKD ++K
Sbjct: 756 GPESDVWSAGVILYILLSGVPPFWAESEPGIFRQILLGKLDFHSEPWPSISDSAKDLIRK 935
Query: 299 LLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASI 358
+L ++P+ RLTA + L HPW+ + A + P+D +VL+ L+QF ++ K+ ALR +A
Sbjct: 936 MLDQNPKTRLTAHEVLRHPWIVDDNIAPDKPLDSAVLSRLKQFSAMNKLKKMALRVIAER 1115
Query: 359 LNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGL 418
L+EEE+ LK+ F ID D +G+I+ +E++ L K + +L ES + +++ A D + G
Sbjct: 1116LSEEEIGGLKELFKMIDTDNSGTITFDELKDGL-KRVGSELMESEIKDLMDAADIDKSGT 1292
Query: 419 VDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG---- 474
+D+ EF+AAT+H+++LE ++ +AF FD D GYIT +E++ G
Sbjct: 1293IDYGEFIAATVHLNKLEREEN------LVSAFSYFDKDGSGYITLDEIQQACKDFGLDDI 1454
Query: 475 SIDPLLEEADIDKDGKISLSEFRRLLRTASMG 506
ID +++E D D DG+I EF ++R + G
Sbjct: 1455HIDDMIKEIDQDNDGQIDYGEFAAMMRKGNGG 1550
>TC206377 similar to UP|O81390 (O81390) Calcium-dependent protein kinase ,
partial (74%)
Length = 1644
Score = 370 bits (951), Expect = e-103
Identities = 183/404 (45%), Positives = 274/404 (67%), Gaps = 6/404 (1%)
Frame = +3
Query: 105 EDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAA 163
ED+K+E+ I++ ++G N+V+F A++D V++VMELC GGEL DRI+A Y+E+ A+
Sbjct: 24 EDMKREIQIMQHLSGQPNIVEFKGAYEDRFSVHLVMELCAGGELFDRIIAQGHYSERAAS 203
Query: 164 VVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIV 223
+ R ++ V CH G++HRD+KPENFL S + + LKATDFGLS FI+ GK +HD+V
Sbjct: 204 SLCRSIVNVVHICHFMGVMHRDLKPENFLLSSKDDHATLKATDFGLSVFIEQGKVYHDMV 383
Query: 224 GSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRK 283
GSAYYVAPEVL+R G + D+WS G+I YILL G PFW +TE GIF +L + DF +
Sbjct: 384 GSAYYVAPEVLRRSYGKEIDIWSAGIILYILLSGVPPFWAETEKGIFNAILEGEIDFVSE 563
Query: 284 PWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVT 343
PWP+IS++AKD V+K+L +DP R+T++Q L HPW+REGG+AS+ PID +VL+ ++QF
Sbjct: 564 PWPSISDSAKDLVRKMLTQDPNKRITSSQVLEHPWMREGGDASDKPIDSAVLSRMKQFRA 743
Query: 344 YSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESR 403
++ K+ AL+ +A L+EEE+ LK F +D D +G+I+ EE++ LA+ + KL E+
Sbjct: 744 MNKLKKLALKVIAENLSEEEIKGLKAMFANMDTDNSGTITYEELKTGLAR-IGSKLSEAE 920
Query: 404 VLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITP 463
V +++ A D + +G +D+ EF++AT+H H+LE + AF+ FD D GYIT
Sbjct: 921 VKQLMDAADVDGNGSIDYLEFISATMHRHRLERDE------HLYKAFQYFDKDNSGYITR 1082
Query: 464 EELRM-----HTGLRGSIDPLLEEADIDKDGKISLSEFRRLLRT 502
+EL + G +I ++ E D D DG+I+ EF ++R+
Sbjct: 1083DELEIAMTQNGMGDEATIKEIISEVDADNDGRINYEEFCAMMRS 1214
>TC224885 similar to UP|Q93YI3 (Q93YI3) Calcium dependent calmodulin
independent protein kinase (Fragment), partial (92%)
Length = 1484
Score = 361 bits (927), Expect = e-100
Identities = 182/386 (47%), Positives = 257/386 (66%), Gaps = 6/386 (1%)
Frame = +2
Query: 122 NVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRG 180
N+V+ A++D V +VMELC GGEL DRI+ Y+E+ AA RQ++ V CH G
Sbjct: 5 NIVELKGAYEDRHSVNLVMELCAGGELFDRIITKGHYSERAAANSCRQIVTVVHNCHSMG 184
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGP 240
++HRD+KPEN L + +DSPLKATDFGLS F KPG F D+VGSAYYVAPEVL+R GP
Sbjct: 185 VMHRDLKPENVLLLNKNDDSPLKATDFGLSVFFKPGDVFRDLVGSAYYVAPEVLRRSYGP 364
Query: 241 QSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL 300
++D+WS GVI YILL G PFW + E GIF +LR DF PWP+IS++AKD VKK+L
Sbjct: 365 EADIWSAGVILYILLSGVPPFWAENEQGIFDAILRGHIDFASDPWPSISSSAKDLVKKML 544
Query: 301 IKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILN 360
DP+ RL+A + L+HPW+R G+A + P+DI+VL ++QF ++ K+ AL+ +A L+
Sbjct: 545 RADPKERLSAVEVLNHPWMRVDGDAPDKPLDIAVLTRMKQFRAMNKLKKVALKVIAENLS 724
Query: 361 EEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVD 420
EEE+ LK+ F ++D D +G+I+ EE++ L K L KL ES V ++++A D + +G +D
Sbjct: 725 EEEIIGLKEMFKSMDTDNSGTITFEELKAGLPK-LGTKLSESEVRQLMEAADVDGNGTID 901
Query: 421 FTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL-----RMHTGLRGS 475
+ EF+ AT+H++++E D AFE FD DK GYIT EEL + + G +
Sbjct: 902 YIEFITATMHMNRMERED------HLYKAFEYFDKDKSGYITMEELESALKKYNMGDEKT 1063
Query: 476 IDPLLEEADIDKDGKISLSEFRRLLR 501
I ++ E D D DG+I+ EF ++R
Sbjct: 1064IKEIIAEVDADNDGRINYDEFVAMMR 1141
>TC211152 UP|Q84P28 (Q84P28) Seed calcium dependent protein kinase b,
complete
Length = 1740
Score = 240 bits (613), Expect(2) = e-100
Identities = 125/294 (42%), Positives = 190/294 (64%), Gaps = 4/294 (1%)
Frame = +1
Query: 212 FIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFK 271
F KPG+ FHD+VGS YYVAPEVL ++ GP+ DVWS GVI YILL G PFW +TE GIF+
Sbjct: 580 FYKPGQAFHDVVGSPYYVAPEVLCKQYGPEVDVWSAGVILYILLSGVPPFWAETEAGIFR 759
Query: 272 EVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPID 331
++L DF +PWP+IS AK+ VK++L +DP+ R++A + L +PWV + A + P+D
Sbjct: 760 QILNGDLDFVSEPWPSISENAKELVKQMLDRDPKKRISAHEVLCNPWVVD-DIAPDKPLD 936
Query: 332 ISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQAL 391
+VL L+ F ++ K+ ALR +A L+EEE+ LK+ F ID D +G+I+ EE+++ L
Sbjct: 937 SAVLTRLKHFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKEGL 1116
Query: 392 AKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFE 451
K + L ES + +++A D + +G +D+ EF+AATLH++++E ++ AAF
Sbjct: 1117-KSVGSNLMESEIKSLMEAADIDNNGSIDYGEFLAATLHLNKMEREEN------LVAAFA 1275
Query: 452 KFDLDKDGYITPEELRMHTGLRG----SIDPLLEEADIDKDGKISLSEFRRLLR 501
FD D GYIT +EL+ +D +++E D D DG+I +EF +++
Sbjct: 1276YFDKDGSGYITIDELQQACKDFSLGDVHLDEMIKEIDQDNDGRIDYAEFAAMMK 1437
Score = 143 bits (360), Expect(2) = e-100
Identities = 69/152 (45%), Positives = 101/152 (66%), Gaps = 1/152 (0%)
Frame = +3
Query: 60 ERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAG 119
+ Y LGK LG GQFG TY+ K +G A K + K K++ +DV +E+ I+ ++
Sbjct: 120 DHYVLGKKLGQGQFGTTYLCTHKVTGKLYACKSIPKRKLMCQEDYDDVWREIQIMHHLSE 299
Query: 120 HENVVQFYNAFDDDSYVYIVMELCEGGELLDRIL-ANRYTEKDAAVVVRQMLKVAAECHL 178
H NVVQ ++D +V++VMELC GGEL DRI+ Y+E++AA +++ ++ V CH
Sbjct: 300 HPNVVQIQGTYEDSVFVHLVMELCAGGELFDRIIQKGHYSEREAAKLIKTIVGVVEACHS 479
Query: 179 RGLVHRDMKPENFLFKSTREDSPLKATDFGLS 210
G++HRD+KPENFLF + ED+ +KATDFGLS
Sbjct: 480 LGVMHRDLKPENFLFDTPGEDAQMKATDFGLS 575
>TC217368 similar to UP|Q93YF4 (Q93YF4) Calcium-dependent protein kinase 2,
partial (66%)
Length = 1349
Score = 345 bits (884), Expect = 4e-95
Identities = 177/372 (47%), Positives = 247/372 (65%), Gaps = 5/372 (1%)
Frame = +1
Query: 139 VMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTR 197
VMELC GGEL DRI+ YTE+ AA + + ++ V CH G++HRD+KPENFLF +
Sbjct: 1 VMELCAGGELFDRIIQRGHYTERQAAELTKTIVGVVEACHSLGVMHRDLKPENFLFINQH 180
Query: 198 EDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCG 257
EDS LK DFGLS F KPG F+D+VGS YYVAPEVL+++ GP++DVWS GVI YILL G
Sbjct: 181 EDSLLKTIDFGLSVFFKPGDIFNDVVGSPYYVAPEVLRKRYGPEADVWSAGVILYILLSG 360
Query: 258 RRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHP 317
PFW + E GIF++VLR DF PWP+IS +AKD V+K+L++DPR RLTA Q L HP
Sbjct: 361 VPPFWAENEQGIFEQVLRGDLDFSSDPWPSISESAKDLVRKMLVRDPRRRLTAHQVLCHP 540
Query: 318 WVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAIDVD 377
W++ G A + P+D +VL+ L+QF ++ K+ AL +A L+EEE+A LK+ F ID D
Sbjct: 541 WIQVDGVAPDKPLDSAVLSRLKQFSAMNKLKKMALIIIAESLSEEEIAGLKEMFKMIDAD 720
Query: 378 KNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEH 437
+G I+ EE++ L K + LKES + +++QA D + G +D+ EF+AATLH +++E
Sbjct: 721 NSGQITFEELKAGL-KRVGANLKESEIYDLMQAADVDNSGTIDYGEFLAATLHRNKIERE 897
Query: 438 DSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRG----SIDPLLEEADIDKDGKISL 493
D+ AAF FD D GYIT EEL+ G ++ +++E D D DG+I
Sbjct: 898 DN------LFAAFSYFDKDGSGYITQEELQQACDEFGIKDVRLEEIIKEIDEDNDGRIDY 1059
Query: 494 SEFRRLLRTASM 505
+EF +++ ++
Sbjct: 1060NEFVAMMQKGNL 1095
>TC224884 similar to UP|Q93YI3 (Q93YI3) Calcium dependent calmodulin
independent protein kinase (Fragment), partial (82%)
Length = 1302
Score = 321 bits (823), Expect = 4e-88
Identities = 161/335 (48%), Positives = 227/335 (67%), Gaps = 5/335 (1%)
Frame = +1
Query: 172 VAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAP 231
V +CH G++HRD+KPENFLF S E+SPLKATDFGLS F KPG F D+VGSAYYVAP
Sbjct: 4 VVHDCHTMGVMHRDLKPENFLFLSKDENSPLKATDFGLSVFFKPGDVFKDLVGSAYYVAP 183
Query: 232 EVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNA 291
EVL+R GP +D+WS GVI +ILL G PFW K E GIF +LR DF PWP+IS++
Sbjct: 184 EVLRRSYGPGADIWSAGVILFILLSGVPPFWSKKEQGIFDAILRGHIDFTSDPWPSISSS 363
Query: 292 AKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFA 351
AKD VKK+L DP+ RL+A + L HPW+RE G AS+ P+D++VL+ ++QF ++ K+ A
Sbjct: 364 AKDLVKKMLRADPKQRLSAVEVLDHPWMREDG-ASDKPLDVAVLSRMKQFRAMNKLKKVA 540
Query: 352 LRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAI 411
L+ +A L+EEE+ LK+ F ++D D +G+I+ EE++ L K L K+ ES V ++++A
Sbjct: 541 LKVIAENLSEEEIIGLKEMFKSMDTDNSGTITFEELKAGLPK-LGTKVSESEVRQLMEAA 717
Query: 412 DSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL----- 466
D + +G +D+ EF+ AT+H++++E D AFE FD D+ GYIT EEL
Sbjct: 718 DVDGNGTIDYIEFITATMHMNRMERED------HLYKAFEYFDKDRSGYITMEELESTLK 879
Query: 467 RMHTGLRGSIDPLLEEADIDKDGKISLSEFRRLLR 501
+ + G +I ++ E D D DG+I+ EF ++R
Sbjct: 880 KYNMGDEKTIKEIIVEVDTDNDGRINYDEFVAMMR 984
>TC209701 similar to UP|Q8LDS1 (Q8LDS1) Calcium-dependent protein kinase,
partial (30%)
Length = 799
Score = 275 bits (703), Expect = 4e-74
Identities = 141/159 (88%), Positives = 150/159 (93%)
Frame = +3
Query: 361 EEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVD 420
EEELAD+KDQFDAIDVDKNGSISLEEMRQALAKDL WKLKESRVLEILQAID+NTDGLVD
Sbjct: 3 EEELADIKDQFDAIDVDKNGSISLEEMRQALAKDLPWKLKESRVLEILQAIDNNTDGLVD 182
Query: 421 FTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRMHTGLRGSIDPLL 480
F EFVAATLHVHQLEE DSDKWQ SQAAFEKFDLDKDGYITPEELRMHT LRGS+DPLL
Sbjct: 183 FREFVAATLHVHQLEE-DSDKWQQLSQAAFEKFDLDKDGYITPEELRMHTCLRGSVDPLL 359
Query: 481 EEADIDKDGKISLSEFRRLLRTASMGSQTVTTPTGYQRR 519
EEADIDKDGKISL EFRRLLRTASMGSQ V++P+ ++R+
Sbjct: 360 EEADIDKDGKISLPEFRRLLRTASMGSQNVSSPSVHRRQ 476
>TC234066 similar to UP|Q8LDS1 (Q8LDS1) Calcium-dependent protein kinase,
partial (28%)
Length = 445
Score = 266 bits (679), Expect = 2e-71
Identities = 134/146 (91%), Positives = 137/146 (93%)
Frame = +1
Query: 315 SHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAI 374
+HPWVREGGEA EIPIDISVLNN+RQFV YSR KQFALRALAS LNE EL+DLKDQFDAI
Sbjct: 7 AHPWVREGGEALEIPIDISVLNNMRQFVKYSRLKQFALRALASTLNEGELSDLKDQFDAI 186
Query: 375 DVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQL 434
DVDKNGSISLEEMRQALAKD WKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQ
Sbjct: 187 DVDKNGSISLEEMRQALAKDQPWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQ* 366
Query: 435 EEHDSDKWQLRSQAAFEKFDLDKDGY 460
EEHDSDKWQ RSQAAFEKFDLDKDGY
Sbjct: 367 EEHDSDKWQQRSQAAFEKFDLDKDGY 444
>TC222915 homologue to UP|Q84P29 (Q84P29) Seed calcium dependent protein
kinase a, partial (45%)
Length = 688
Score = 243 bits (621), Expect = 1e-64
Identities = 114/227 (50%), Positives = 165/227 (72%)
Frame = +1
Query: 176 CHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLK 235
CH G++HRD+KPENFLF + E + LK TDFGLS F KPG+ F D+VGS YYVAPEVL+
Sbjct: 7 CHSLGVMHRDLKPENFLFDTVEEGAKLKTTDFGLSVFYKPGETFCDVVGSPYYVAPEVLR 186
Query: 236 RKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDF 295
+ GP++DVWS GVI YILL G PFW +TE GIF+++L + DF+ +PWP+IS++AKD
Sbjct: 187 KHYGPEADVWSAGVILYILLSGVPPFWAETEQGIFRQILLGRIDFQSEPWPSISDSAKDL 366
Query: 296 VKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRAL 355
++K+L ++P+ R+TA Q L HPW+ + A + P+D +VL+ L+QF ++ K+ ALR +
Sbjct: 367 IRKMLDRNPKTRVTAHQVLCHPWIVDDNIAPDKPLDSAVLSRLKQFSAMNKLKKMALRVI 546
Query: 356 ASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKES 402
A L+EEE+ LK+ F ID D +G+I+ +E+++ L K + +L ES
Sbjct: 547 AERLSEEEIGGLKELFRMIDADNSGTITFDELKEGL-KRVGSELMES 684
>TC225062 similar to UP|Q8W4I7 (Q8W4I7) Calcium-dependent protein kinase,
partial (60%)
Length = 1701
Score = 243 bits (619), Expect = 2e-64
Identities = 125/290 (43%), Positives = 182/290 (62%), Gaps = 6/290 (2%)
Frame = +1
Query: 221 DIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDF 280
+IVGS YY+APEVLKR GP+ D+WS GVI YILLCG PFW ++E G+ + +LR DF
Sbjct: 4 EIVGSPYYMAPEVLKRNYGPEIDIWSAGVILYILLCGVPPFWAESEQGVAQAILRGLIDF 183
Query: 281 RRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQ 340
+R+PWP+IS +AK V+++L DP+ RLTA Q L HPW++ +A +P+ V + L+Q
Sbjct: 184 KREPWPSISESAKSLVRQMLEPDPKLRLTAKQVLEHPWIQNAKKAPNVPLGDVVKSRLKQ 363
Query: 341 FVTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLK 400
F +RFK+ ALR +A L+ EE+ D+KD F +D D +G +S+EE++ ++ L
Sbjct: 364 FSMMNRFKRKALRVIADFLSNEEVEDIKDMFKKMDNDNDGIVSIEELKAGF-RNFGSLLA 540
Query: 401 ESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGY 460
+S V +++A+DSN G +D+ EFVA +LH+ ++ D AF FD D +GY
Sbjct: 541 DSEVQLLIEAVDSNGKGTLDYGEFVAVSLHLRRMANDD------HLHKAFSYFDKDGNGY 702
Query: 461 ITPEELR---MHTGLRGSID---PLLEEADIDKDGKISLSEFRRLLRTAS 504
I P+ELR M G D + E D DKDG+IS EF +++T +
Sbjct: 703 IEPDELRNALMEDGADDCTDVANDIFLEVDTDKDGRISYDEFVAMMKTGT 852
>TC208216 similar to UP|O48565 (O48565) Calcium-dependent protein kinase,
partial (42%)
Length = 1316
Score = 235 bits (599), Expect = 4e-62
Identities = 109/208 (52%), Positives = 152/208 (72%), Gaps = 1/208 (0%)
Frame = +3
Query: 56 KDFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILK 115
++ + RY LG+ LG G+FG TY+ DK +G+ +A K + K K+ +EDV++EV I++
Sbjct: 435 REIEARYELGRELGRGEFGITYLCTDKGTGEELACKSISKKKLRTAIDIEDVRREVEIMR 614
Query: 116 AVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAA 174
+ H N+V + ++DD+ V++VMELCEGGEL DRI+A YTE+ AA V + +++V
Sbjct: 615 HLPQHANIVTLKDTYEDDNAVHLVMELCEGGELFDRIVARGHYTERAAAAVTKTIVEVVQ 794
Query: 175 ECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVL 234
CH +G++HRD+KPENFLF + +E + LKA DFGLS F KPG+KF++IVGS YY+APEVL
Sbjct: 795 MCHKQGVMHRDLKPENFLFANKKETAALKAIDFGLSVFFKPGEKFNEIVGSPYYMAPEVL 974
Query: 235 KRKSGPQSDVWSIGVITYILLCGRRPFW 262
KR GP+ D+WS GVI YILLCG PFW
Sbjct: 975 KRNYGPEVDIWSAGVILYILLCGVPPFW 1058
>TC219157 similar to UP|Q9LET1 (Q9LET1) Calcium-dependent protein
kinase-like, partial (33%)
Length = 595
Score = 223 bits (569), Expect = 1e-58
Identities = 109/197 (55%), Positives = 144/197 (72%)
Frame = +3
Query: 178 LRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRK 237
L+G+VHRD+KPENFL+ S E+S LK DFGLSD++KP ++ +DIVGSAYYVAPEVL R
Sbjct: 3 LQGVVHRDLKPENFLYISKDENSTLKVIDFGLSDYVKPDERLNDIVGSAYYVAPEVLHRS 182
Query: 238 SGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVK 297
G ++D+WSIGVI YILLCG RPFW +TE GIF+ VL+ P F PWP++S AKDFVK
Sbjct: 183 YGTEADMWSIGVIAYILLCGSRPFWARTESGIFRAVLKADPSFEEAPWPSLSADAKDFVK 362
Query: 298 KLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALAS 357
+LL KD R RLTAAQALSHPW+ + +IP D+ + ++ ++ S ++ AL ALA
Sbjct: 363 RLLNKDYRKRLTAAQALSHPWLVNHCDDVKIPFDMIIHKLVKTYICSSSLRKSALGALAK 542
Query: 358 ILNEEELADLKDQFDAI 374
L +LA L++QF+ +
Sbjct: 543 TLTLVQLAYLREQFNML 593
>TC208071 UP|Q8GRZ5 (Q8GRZ5) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1188
Score = 186 bits (473), Expect = 2e-47
Identities = 97/275 (35%), Positives = 159/275 (57%), Gaps = 1/275 (0%)
Frame = +3
Query: 62 YSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHE 121
Y + + +G G+FG + S A K ++KS ++ ++ E + ++ H
Sbjct: 36 YEVSEEIGRGRFGTIFRCFHPLSNQPYACKLIDKSLLLDSTDRHCLQNEPKFMSLLSPHP 215
Query: 122 NVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAAVVVRQMLKVAAECHLRGL 181
N++Q ++ F+DD Y+ IVM+LC+ L DR+L ++E AA +++ +L+ A CH G+
Sbjct: 216 NILQIFHVFEDDHYLSIVMDLCQPHTLFDRMLHAPFSESQAASLIKNLLEAVAHCHRLGV 395
Query: 182 VHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEV-LKRKSGP 240
HRD+KP+N LF S LK DFG +++ G+ +VG+ YYVAPEV L R+
Sbjct: 396 AHRDIKPDNILFDSA---DNLKLADFGSAEWFGDGRSMSGVVGTPYYVAPEVLLGREYDE 566
Query: 241 QSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL 300
+ DVWS GVI YI+L G PF+ + IF+ V++ F + + T+S AAKD ++K++
Sbjct: 567 KVDVWSCGVILYIMLAGIPPFYGDSAAEIFEAVVKANLRFPSRIFRTVSPAAKDLLRKMI 746
Query: 301 IKDPRARLTAAQALSHPWVREGGEASEIPIDISVL 335
+D R +A QAL HPW+ G+ +E+ + S +
Sbjct: 747 SRDSSRRFSAEQALRHPWILSAGDTAELT*ECSYM 851
>TC217983 UP|Q8GSL0 (Q8GSL0) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1203
Score = 186 bits (472), Expect = 2e-47
Identities = 97/268 (36%), Positives = 155/268 (57%), Gaps = 1/268 (0%)
Frame = +3
Query: 62 YSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHE 121
Y L + +G G+FG + S + A K ++KS + + ++ E + ++ H
Sbjct: 60 YQLSEEIGRGRFGTIFRCFHPLSNEPYACKLIDKSLLHDSTDRDCLQNEPKFMTLLSPHP 239
Query: 122 NVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAAVVVRQMLKVAAECHLRGL 181
N++Q ++ F+DD Y+ IVM+LC+ L DR++ E AA +++ +L+ A CH G+
Sbjct: 240 NILQIFHVFEDDQYLSIVMDLCQPHTLFDRMVDGPIQESQAAALMKNLLEAVAHCHRLGV 419
Query: 182 VHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEV-LKRKSGP 240
HRD+KP+N LF S LK DFG +++ G+ +VG+ YYVAPEV L R+
Sbjct: 420 AHRDIKPDNILFDSA---DNLKLADFGSAEWFGDGRSMSGVVGTPYYVAPEVLLGREYDE 590
Query: 241 QSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL 300
+ DVWS GVI YI+L G PF+ + IF+ V+R F + + T+S AAKD ++K++
Sbjct: 591 KVDVWSCGVILYIMLAGIPPFYGDSAAEIFEAVVRANLRFPSRIFRTVSPAAKDLLRKMI 770
Query: 301 IKDPRARLTAAQALSHPWVREGGEASEI 328
+D R +A QAL HPW+ G+ +E+
Sbjct: 771 CRDSSRRFSAEQALRHPWILSAGDTAEL 854
>TC207315 UP|Q8H1P2 (Q8H1P2) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1063
Score = 181 bits (458), Expect = 9e-46
Identities = 102/265 (38%), Positives = 155/265 (58%), Gaps = 2/265 (0%)
Frame = +3
Query: 60 ERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAG 119
E Y + + LG G+FG + + S A K +EK ++ L ++ E + ++
Sbjct: 51 EEYQVLEELGRGRFGTVFRCFHRTSNKFYAAKLIEKRRL-LNEDRRCIEMEAKAMSFLSP 227
Query: 120 HENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANR-YTEKDAAVVVRQMLKVAAECHL 178
H N++Q +AF+D IV+ELC+ LLDRI A TE AA +++Q+L+ A CH
Sbjct: 228 HPNILQIMDAFEDADSCSIVLELCQPHTLLDRIAAQGPLTEPHAASLLKQLLEAVAHCHA 407
Query: 179 RGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLK-RK 237
+GL HRD+KPEN LF E + LK +DFG ++++ G +VG+ YYVAPEV+ R+
Sbjct: 408 QGLAHRDIKPENILFD---EGNKLKLSDFGSAEWLGEGSSMSGVVGTPYYVAPEVIMGRE 578
Query: 238 SGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVK 297
+ DVWS GVI Y +L G PF+ ++ IF+ VLR F + ++S AKD ++
Sbjct: 579 YDEKVDVWSSGVILYAMLAGFPPFYGESAPEIFESVLRANLRFPSLIFSSVSAPAKDLLR 758
Query: 298 KLLIKDPRARLTAAQALSHPWVREG 322
K++ +DP R++A QAL HPW+ G
Sbjct: 759 KMISRDPSNRISAHQALRHPWILTG 833
>TC225868 UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, complete
Length = 2261
Score = 181 bits (458), Expect = 9e-46
Identities = 112/356 (31%), Positives = 187/356 (52%), Gaps = 5/356 (1%)
Frame = +2
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+Y LG+LLGHG F Y +G VA+K + K K+V +E +K+E++ + V H
Sbjct: 485 KYELGRLLGHGTFAKVYHARHLKTGKSVAMKVVGKEKVVKVGMMEQIKREISAMNMVK-H 661
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAAVVVRQMLKVAAECHLRG 180
N+VQ + S +YI MEL GGEL ++I R E+ A + +Q++ CH RG
Sbjct: 662 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIARGRLREEMARLYFQQLISAVDFCHSRG 841
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLSDF---IKPGKKFHDIVGSAYYVAPEVLKRK 237
+ HRD+KPEN L +D LK TDFGLS F ++ H G+ YVAPEV+ ++
Sbjct: 842 VYHRDLKPENLLLD---DDGNLKVTDFGLSTFSEHLRHDGLLHTTCGTPAYVAPEVIGKR 1012
Query: 238 --SGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDF 295
G ++D+WS GVI Y+LL G PF D ++K++ R DF+ PW S+ A+
Sbjct: 1013GYDGAKADIWSCGVILYVLLAGFLPFQDDNLVALYKKIYRG--DFKCPPW--FSSEARRL 1180
Query: 296 VKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRAL 355
+ KLL +P R+T ++ + W ++ P+ +++ R+ + + + +
Sbjct: 1181ITKLLDPNPNTRITISKIMDSSWFKK-------PVPKNLMGKKREELDLEEKIKQHEQEV 1339
Query: 356 ASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAI 411
++ +N + L + FD + + +E+R A + S + SR+ ++ +A+
Sbjct: 1340STTMNAFHIISLSEGFDLSPLFEEKKREEKELRFATTRPASSVI--SRLEDLAKAV 1501
>TC205579 homologue to UP|Q43676 (Q43676) Calcium dependent protein kinase ,
partial (52%)
Length = 1481
Score = 171 bits (433), Expect = 7e-43
Identities = 91/238 (38%), Positives = 143/238 (59%), Gaps = 4/238 (1%)
Frame = +1
Query: 273 VLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDI 332
VL+ DF PWP IS++AKD ++K+L P RLTA Q L HPW+ E G A + +D
Sbjct: 4 VLKGLIDFDSDPWPLISDSAKDLIRKMLCSRPSERLTAHQVLCHPWICENGVAPDRSLDP 183
Query: 333 SVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLEEMRQALA 392
+VL+ L+QF ++ K+ ALR +A L+EEE+A L++ F A+D D +G+I+ +E++ L
Sbjct: 184 AVLSRLKQFSAMNKLKKMALRVIAESLSEEEIAGLREMFQAMDTDNSGAITFDELKAGLR 363
Query: 393 KDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAATLHVHQLEEHDSDKWQLRSQAAFEK 452
+ S LK+ + ++++A D + G +D+ EF+AAT+H+++LE + AAF+
Sbjct: 364 RYGS-TLKDIEIRDLMEAADVDKSGTIDYGEFIAATVHLNKLEREE------HLIAAFQY 522
Query: 453 FDLDKDGYITPEELRMHTGLRGSIDPLLE----EADIDKDGKISLSEFRRLLRTASMG 506
FD D GYIT +EL+ + D LE E D D DG+I EF +++ + G
Sbjct: 523 FDKDGSGYITVDELQQACAEQNMTDAFLEDIIREVDQDNDGRIDYGEFAAMMQKGNAG 696
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,758,695
Number of Sequences: 63676
Number of extensions: 239377
Number of successful extensions: 2359
Number of sequences better than 10.0: 591
Number of HSP's better than 10.0 without gapping: 1997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2048
length of query: 520
length of database: 12,639,632
effective HSP length: 102
effective length of query: 418
effective length of database: 6,144,680
effective search space: 2568476240
effective search space used: 2568476240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0119.8


