

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118c.15
(602 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
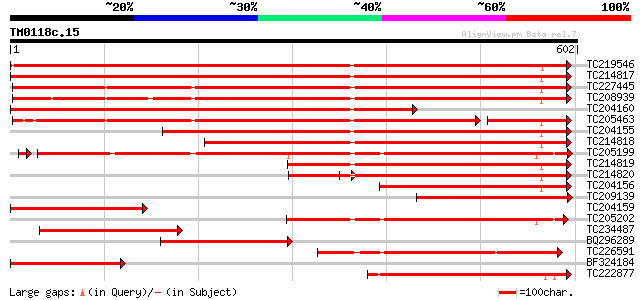
Score E
Sequences producing significant alignments: (bits) Value
TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete 825 0.0
TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, ... 801 0.0
TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum H... 637 0.0
TC208939 UP|Q39830 (Q39830) BiP isoform A, complete 614 e-176
TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate ... 608 e-174
TC205463 UP|Q39804 (Q39804) BiP isoform B, complete 530 e-169
TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, ... 560 e-160
TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous hea... 492 e-139
TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 459 e-131
TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat sh... 372 e-103
TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight h... 296 1e-80
TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 249 3e-66
TC209139 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein... 227 1e-59
TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial ... 224 1e-58
TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa pro... 215 5e-56
TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%) 211 6e-55
BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cyto... 200 1e-51
TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock ... 191 8e-49
BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chapero... 189 4e-48
TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa p... 175 6e-44
>TC219546 UP|HS70_SOYBN (P26413) Heat shock 70 kDa protein, complete
Length = 2099
Score = 825 bits (2132), Expect = 0.0
Identities = 422/610 (69%), Positives = 503/610 (82%), Gaps = 14/610 (2%)
Frame = +1
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MAT K+ KAIGIDLGTTYSCV VW ++RVEIIPN+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 1 MAT-KEGKAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKN 177
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ DMKLWPFKV DKPMI V YKGEE+K +A
Sbjct: 178 QVAMNPQNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCDKPMIVVNYKGEEKKFSA 357
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KM+EVAEA+LGH VK AV+TVPAYFN++QRQ+TKDAG I+G +V+RIINEP
Sbjct: 358 EEISSMVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEP 537
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK RKGEQNVL+FDLGGGTFDVS++TI+EGIF+V AT GDT+LGG DFD
Sbjct: 538 TAAAIAYGLDKKASRKGEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDFD 717
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTTIE+DSL GID +
Sbjct: 718 NRMVNHFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFY 897
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMF 360
T+T+ FE++N DLF KCME VEKCL +AKI KS+VHE+VLVGGSTRIPKV QLL++ F
Sbjct: 898 ATITRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLLQDFF 1077
Query: 361 SVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVM 420
+ KELCKSINPDEAVAYGAAVQAAILS +GD+KV++LLLLDV PLSLG+E GVM
Sbjct: 1078N----GKELCKSINPDEAVAYGAAVQAAILSGQGDEKVQDLLLLDVTPLSLGLETAGGVM 1245
Query: 421 SVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKG 480
+VLIP+NTTIPTKKE+ F T SDNQ V I+V+EGE A+T N LGKFEL+G PAP+G
Sbjct: 1246TVLIPRNTTIPTKKEQIFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELTGIPPAPRG 1425
Query: 481 DTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDE 540
+INVCFD+DA+GI+ VSAED + +K ITITN GRLS+EE+ +MV+DA +YKAEDE
Sbjct: 1426VPQINVCFDIDANGILNVSAEDKTAGVKNKITITNDKGRLSKEEIEKMVKDAERYKAEDE 1605
Query: 541 EVRKKVKAKNSLENYAYEARDRVK--------------KLEKMVEEVIEWLDRNQLAEAE 586
EV+KKV+AKNSLENYAY R+ +K K+EK VE+ I+WL+ NQ+AE +
Sbjct: 1606EVKKKVEAKNSLENYAYNMRNTIKDEKIGGKLSPDEKQKIEKAVEDAIQWLEGNQMAEVD 1785
Query: 587 EFEYKKQELE 596
EFE K++ELE
Sbjct: 1786EFEDKQKELE 1815
>TC214817 homologue to UP|Q9M4E7 (Q9M4E7) Heat shock protein 70, complete
Length = 2364
Score = 801 bits (2069), Expect = 0.0
Identities = 413/610 (67%), Positives = 491/610 (79%), Gaps = 14/610 (2%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYV FTDTERL+GDAA N
Sbjct: 86 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKN 265
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ D+KLWPFKV+ G DKPMI V YKGEE++ AA
Sbjct: 266 QVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIPGAADKPMIVVNYKGEEKQFAA 445
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KM+E+AEAYLG VK AV+TVPAYFN++QRQ+TKDAG IAG +VMRIINEP
Sbjct: 446 EEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEP 625
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGIF+V AT GDT+LGG DFD
Sbjct: 626 TAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 805
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTTIE+DSL GID +
Sbjct: 806 NRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFY 985
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMF 360
TVT+ FE+LN DLF KCME VEKCL +AK+ K V ++VLVGGSTRIPKVQQLL++ F
Sbjct: 986 STVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQLLQDFF 1165
Query: 361 SVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVM 420
+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLSLG+E GVM
Sbjct: 1166N----GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVM 1333
Query: 421 SVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKG 480
+VLIP+NTTIPTKKE+ F T SDNQ V I+V+EGE A+T N LGKFELSG PAP+G
Sbjct: 1334TVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTRDNNLLGKFELSGIPPAPRG 1513
Query: 481 DTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDE 540
+I VCFD+DA+GI+ VSAED + K ITITN GRLS+E++ +MV++A +YK+EDE
Sbjct: 1514VPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDE 1693
Query: 541 EVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEWLDRNQLAEAE 586
E +KKV+AKN+LENYAY R+ V KK+E +E+ I+WLD NQLAEA+
Sbjct: 1694EHKKKVEAKNALENYAYNMRNTVKDDKIGEKLDPADKKKIEDAIEQAIQWLDSNQLAEAD 1873
Query: 587 EFEYKKQELE 596
EFE K +ELE
Sbjct: 1874EFEDKMKELE 1903
>TC227445 homologue to UP|O22639 (O22639) Endoplasmic reticulum HSC70-cognate
binding protein precursor, complete
Length = 2254
Score = 637 bits (1643), Expect = 0.0
Identities = 335/609 (55%), Positives = 441/609 (72%), Gaps = 16/609 (2%)
Frame = +2
Query: 4 SKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLA 63
+K IGIDLGTTYSCV V+ + VEII N+ GNR TPS+VAFTD+ERL+G+AA N A
Sbjct: 200 TKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVAFTDSERLIGEAAKNLAA 379
Query: 64 LNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYK-GEERKLAAEE 122
+NP T+FD KRLIGR+F D+ VQ+DMKL P+K+V KP I V K GE + + EE
Sbjct: 380 VNPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKD-GKPYIQVKIKDGETKVFSPEE 556
Query: 123 ISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTA 182
IS+M+L KMKE AEA+LG ++ AV+TVPAYFN+AQRQ+TKDAG IAG +V RIINEPTA
Sbjct: 557 ISAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTA 736
Query: 183 ASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNN 242
A+IAYGLDKKG GE+N+LVFDLGGGTFDVS++TID G+F+V AT GDT+LGG DFD
Sbjct: 737 AAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQR 907
Query: 243 LVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVT 302
++ + + ++KH KDI+++ ++L +LR E+AKR LSS Q +E++SL G+D
Sbjct: 908 IMEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEP 1087
Query: 303 VTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSV 362
+T+ FE+LN DLF K M V+K + +A + KS++ E+VLVGGSTRIPKVQQLLK+ F
Sbjct: 1088LTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFDG 1267
Query: 363 HGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSV 422
KE K +NPDEAVAYGAAVQ +ILS EG ++ +++LLLDV PL+LGIE V GVM+
Sbjct: 1268----KEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTK 1435
Query: 423 LIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDT 482
LIP+NT IPTKK + F T D QT+V I+V+EGE + T LGKF+LSG PAP+G
Sbjct: 1436LIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTP 1615
Query: 483 EINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEV 542
+I V F+VDA+GI+ V AED + ITITN GRLSQEE+ RMVR+A ++ ED++V
Sbjct: 1616QIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIDRMVREAEEFAEEDKKV 1795
Query: 543 RKKVKAKNSLENYAYEARDRV---------------KKLEKMVEEVIEWLDRNQLAEAEE 587
++++ A+NSLE Y Y +++V +K+E V+E +EWLD NQ E E+
Sbjct: 1796KERIDARNSLETYVYNMKNQVSDKDKLADKLESDEKEKIETAVKEALEWLDDNQSVEKED 1975
Query: 588 FEYKKQELE 596
+E K +E+E
Sbjct: 1976YEEKLKEVE 2002
>TC208939 UP|Q39830 (Q39830) BiP isoform A, complete
Length = 2452
Score = 614 bits (1584), Expect = e-176
Identities = 329/609 (54%), Positives = 433/609 (71%), Gaps = 16/609 (2%)
Frame = +3
Query: 4 SKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLA 63
+K IGIDLGTTYSCV V+ + VEII N+ GNR TPS+ +FTD+ERL+G+AA N A
Sbjct: 162 TKLGTVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSW-SFTDSERLIGEAAKNLAA 338
Query: 64 LNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYK-GEERKLAAEE 122
+NP +FD KRLIGR+F D+ VQ+DMKL P+K+V KP I K GE + + EE
Sbjct: 339 VNPERVIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKD-GKPYIQEKIKDGETKVFSPEE 515
Query: 123 ISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTA 182
IS+M+L KMKE AEA+LG ++ AV AYFN+AQRQ+TKDAG IAG +V RIINEPTA
Sbjct: 516 ISAMILTKMKETAEAFLGKKINDAV----AYFNDAQRQATKDAGVIAGLNVARIINEPTA 683
Query: 183 ASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNN 242
A+IAYGLDKKG GE+N+LVFDLGGGTFDVS++TID G+F+V AT GDT+LGG DFD
Sbjct: 684 AAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQR 854
Query: 243 LVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVT 302
++ + + +KH KDI+++ ++L +LR E+AKR LSS Q +E++SL G+D
Sbjct: 855 IMEYFIKLINKKHKKDISKDSRALSKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEP 1034
Query: 303 VTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSV 362
+T+ FE+LN DLF K M V+K + +A + K+++ E+VLVGGSTRIPKVQQLLK+ F
Sbjct: 1035LTRARFEELNNDLFRKTMGPVKKAMEDAGLQKNQIDEIVLVGGSTRIPKVQQLLKDYFDG 1214
Query: 363 HGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSV 422
KE K +NPDEAVAYGAAVQ +ILS EG ++ +++LLLDV PL+LGIE V GVM+
Sbjct: 1215----KEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTK 1382
Query: 423 LIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDT 482
LIP+NT IPTKK + F T D QT+V I+V+EGE + T LGKFELSG PAP+G
Sbjct: 1383LIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFELSGIPPAPRGTP 1562
Query: 483 EINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEV 542
+I V F+VDA+GI+ V AED + ITITN GRLSQEE+ RMVR+A ++ ED++V
Sbjct: 1563QIEVTFEVDANGILNVKAEDKGTGKSEKITITNEKGRLSQEEIERMVREAEEFAEEDKKV 1742
Query: 543 RKKVKAKNSLENYAYEARDRV---------------KKLEKMVEEVIEWLDRNQLAEAEE 587
++++ A+NSLE Y Y +++V +K+E V+E +EWLD NQ E EE
Sbjct: 1743KERIDARNSLETYVYNMKNQVSDKDKLADKLESDEKEKVETAVKEALEWLDDNQSVEKEE 1922
Query: 588 FEYKKQELE 596
+E K +E+E
Sbjct: 1923YEEKLKEVE 1949
>TC204160 homologue to UP|HS72_LYCES (P27322) Heat shock cognate 70 kDa
protein 2, partial (66%)
Length = 1390
Score = 608 bits (1567), Expect = e-174
Identities = 308/433 (71%), Positives = 362/433 (83%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 95 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 274
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ DMKLWPFKV+ G +KPMI V YKGEE++ +A
Sbjct: 275 QVAMNPTNTVFDAKRLIGRRFSDASVQGDMKLWPFKVIPGPAEKPMIVVNYKGEEKQFSA 454
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEP 180
EEISSMVL KMKE+AEAYLG +K AV+TVPAYFN++QRQ+TKDAG I+G +VMRIINEP
Sbjct: 455 EEISSMVLMKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEP 634
Query: 181 TAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFD 240
TAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGIF+V AT GDT+LGG DFD
Sbjct: 635 TAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 814
Query: 241 NNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLH 300
N +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTTIE+DSL GID +
Sbjct: 815 NRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFY 994
Query: 301 VTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMF 360
T+T+ FE+LN DLF KCME VEKCL +AK+ KS VH++VLVGGSTRIPKVQQLL++ F
Sbjct: 995 TTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFF 1174
Query: 361 SVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVM 420
+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV LS +E GVM
Sbjct: 1175N----GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTTLSTALETARGVM 1342
Query: 421 SVLIPKNTTIPTK 433
+VL+ NTTI T+
Sbjct: 1343TVLMTSNTTISTE 1381
>TC205463 UP|Q39804 (Q39804) BiP isoform B, complete
Length = 2383
Score = 530 bits (1365), Expect(2) = e-169
Identities = 287/499 (57%), Positives = 369/499 (73%), Gaps = 2/499 (0%)
Frame = +3
Query: 4 SKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLA 63
+K IGIDL TTYSCV V + VEII N+ GNR TPS+VAFTD+ERL+G+AA A
Sbjct: 183 TKLGTVIGIDL-TTYSCVGVTD--AVEIIANDQGNRITPSWVAFTDSERLIGEAAKIVAA 353
Query: 64 LNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYK-GEERKLAAEE 122
+NP T+FD KRLIGR+F D+ VQ+DMKL P+K+V KP I V K GE + + EE
Sbjct: 354 VNPVRTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKD-GKPYIQVKIKDGETKVFSPEE 530
Query: 123 ISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTA 182
IS+M+L KMKE AEA+LG ++ AV+TVPAYFN+AQRQ+TKDAG IAG +V RIINEPTA
Sbjct: 531 ISAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTA 710
Query: 183 ASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNN 242
A+IAYGLDKKG GE+N+LVFDLGGGTFDVS++TID G+F+V AT GDT+LGG DFD
Sbjct: 711 AAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDTHLGGEDFDQR 881
Query: 243 LVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVT 302
++ + + ++KH KDI+++ ++L +LR E+AKR LSS Q +E++SL G+D
Sbjct: 882 IMEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQVRVEIESLFDGVDFSEP 1061
Query: 303 VTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSV 362
+T+ FE+LN DLF K M V+K + +A + KS++ E+VLVGGSTRIPKVQQLLK+ F
Sbjct: 1062LTRARFEELNNDLFRKTMGPVKKAMEDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDYFD- 1238
Query: 363 HGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSV 422
KE K +NPDEAVAYGAAVQ +ILS EG ++ +++LLLDV PL+LGIE V GVM+
Sbjct: 1239---GKEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILLLDVAPLTLGIETVGGVMTK 1409
Query: 423 LIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDT 482
LIP+NT IPTKK + F T D QT+V I+V+EGE + T LGKF+LSG PAP+G
Sbjct: 1410LIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLSGIPPAPRGTP 1589
Query: 483 EINVCFDVDAD-GIVEVSA 500
+I V F+VDA+ GI+ V A
Sbjct: 1590QIEVTFEVDAERGILNVKA 1646
Score = 84.3 bits (207), Expect(2) = e-169
Identities = 42/104 (40%), Positives = 66/104 (63%), Gaps = 15/104 (14%)
Frame = +1
Query: 508 KKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV---- 563
+K ITN GRLSQEE+ RMVR+A ++ ED++V++++ A+NSLE Y Y ++++
Sbjct: 1672 QKRSPITNEKGRLSQEEIERMVREAEEFAEEDKKVKERIDARNSLETYVYNMKNQISDKD 1851
Query: 564 -----------KKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELE 596
+K+E V+E +EWLD NQ E E++E K +E+E
Sbjct: 1852 KLADKLESDEKEKIETAVKEALEWLDDNQSMEKEDYEEKLKEVE 1983
>TC204155 homologue to UP|Q84QJ3 (Q84QJ3) Heat shock protein 70, partial
(75%)
Length = 1640
Score = 560 bits (1444), Expect = e-160
Identities = 290/448 (64%), Positives = 353/448 (78%), Gaps = 14/448 (3%)
Frame = +2
Query: 163 KDAGEIAGFDVMRIINEPTAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGI 222
KDAG I+G +VMRIINEPTAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGI
Sbjct: 2 KDAGVISGLNVMRIINEPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGI 181
Query: 223 FKVNATVGDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSS 282
F+V AT GDT+LGG DFDN +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSS
Sbjct: 182 FEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACERAKRTLSS 361
Query: 283 TSQTTIELDSLCGGIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVL 342
T+QTTIE+DSL GID + T+T+ FE+LN DLF KCME VEKCL +AK+ KS VH++VL
Sbjct: 362 TAQTTIEIDSLYEGIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVL 541
Query: 343 VGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELL 402
VGGSTRIPKVQQLL++ F+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LL
Sbjct: 542 VGGSTRIPKVQQLLQDFFN----GKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLL 709
Query: 403 LLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDK 462
LLDV PLS G+E GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE +T
Sbjct: 710 LLDVTPLSTGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRD 889
Query: 463 NFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQ 522
N LGKFELSG PAP+G +I VCFD+DA+GI+ VSAED + K ITITN GRLS+
Sbjct: 890 NNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSK 1069
Query: 523 EEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV--------------KKLEK 568
EE+ +MV++A +YK+EDEE +KKV+AKN+LENYAY R+ + KK+E
Sbjct: 1070EEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIKDDKIASKLSSDDKKKIED 1249
Query: 569 MVEEVIEWLDRNQLAEAEEFEYKKQELE 596
+E+ I+WLD NQLAEA+EFE K +ELE
Sbjct: 1250AIEQAIQWLDGNQLAEADEFEDKMKELE 1333
>TC214818 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (68%)
Length = 1554
Score = 492 bits (1266), Expect = e-139
Identities = 255/404 (63%), Positives = 314/404 (77%), Gaps = 14/404 (3%)
Frame = +2
Query: 207 GGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSL 266
GGGTFDVSL+TI+EGIF+V AT GDT+LGG DFDN +VN V F+RK+ KDI+ N ++L
Sbjct: 8 GGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRAL 187
Query: 267 RRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVTVTQGLFEKLNKDLFEKCMEIVEKC 326
RRLR ACE+AKR LSST+QTTIE+DSL GID + T+T+ FE+LN DLF KCME VEKC
Sbjct: 188 RRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRARFEELNMDLFRKCMEPVEKC 367
Query: 327 LVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQ 386
L +AK+ K VH++VLVGGSTRIPKVQQLL++ F+ KELCKSINPDEAVAYGAAVQ
Sbjct: 368 LRDAKMDKRTVHDVVLVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGAAVQ 535
Query: 387 AAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQT 446
AAILS EG++KV++LLLLDV PLSLG+E GVM+VLIP+NTTIPTKKE+ F T SDNQ
Sbjct: 536 AAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQP 715
Query: 447 SVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLR 506
V I+VYEGE +T N LGKFELSG PAP+G +I VCFD+DA+GI+ VSAED +
Sbjct: 716 GVLIQVYEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTG 895
Query: 507 LKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV--- 563
K ITITN GRLS+EE+ +MV++A +YK+EDEE +KKV+AKN+LENY+Y R+ +
Sbjct: 896 QKNKITITNDKGRLSKEEIEKMVQEAEKYKSEDEEHKKKVEAKNALENYSYNMRNTIKDE 1075
Query: 564 -----------KKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELE 596
KK+E +E+ I+WLD NQL EA+EFE K +ELE
Sbjct: 1076KIGGKLDPADKKKIEDAIEQAIQWLDSNQLGEADEFEDKMKELE 1207
>TC205199 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (94%)
Length = 2204
Score = 459 bits (1180), Expect(2) = e-131
Identities = 270/583 (46%), Positives = 372/583 (63%), Gaps = 15/583 (2%)
Frame = +3
Query: 30 EIIPNELGNRTTPSYVAFTD-TERLMGDAAINQLALNPHNTVFDAKRLIGRRFSDRSVQQ 88
++I N G RTTPS VAF E L+G A Q NP NT+F KRLIGRRF D Q+
Sbjct: 186 KVIENSEGARTTPSVVAFNQKAELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDSQTQK 365
Query: 89 DMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVLFKMKEVAEAYLGHEVKRAVI 148
+MK+ P+K+V+ + + ++ + ++ + VL KMKE AE+YLG V +AVI
Sbjct: 366 EMKMVPYKIVKAPNGDAWV----EANGQQYSPSQVGAFVLTKMKETAESYLGKSVSKAVI 533
Query: 149 TVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASIAYGLDKKGWRKGEQNVLVFDLGG 208
TVPAYFN+AQRQ+TKDAG IAG DV RIINEPTAA+++YG++ K E + VFDLGG
Sbjct: 534 TVPAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNK-----EGLIAVFDLGG 698
Query: 209 GTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRR 268
GTFDVS++ I G+F+V AT GDT+LGG DFDN L++FLV+ F+R N D++++ +L+R
Sbjct: 699 GTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTENIDLSKDKLALQR 878
Query: 269 LRLACEKAKRILSSTSQTTIELDSLC----GGIDLHVTVTQGLFEKLNKDLFEKCMEIVE 324
LR A EKAK LSSTSQT I L + G L++T+T+ FE L L E+ +
Sbjct: 879 LREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKAPCK 1058
Query: 325 KCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAA 384
CL +A + EV E++LVGG TR+PKVQ+++ +F K K +NPDEAVA GAA
Sbjct: 1059SCLKDANVSIKEVDEVLLVGGMTRVPKVQEVVSAIFG-----KSPSKGVNPDEAVAMGAA 1223
Query: 385 VQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDN 444
+Q IL + V+ELLLLDV PLSLGIE + G+ + LI +NTTIPTKK + F T +DN
Sbjct: 1224IQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADN 1391
Query: 445 QTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMS 504
QT V IKV +GE N LG+FEL G PAP+G +I V FD+DA+GIV VSA+D S
Sbjct: 1392QTQVGIKVLQGEREMAVDNKSLGEFELVGIPPAPRGMPQIEVTFDIDANGIVTVSAKDKS 1571
Query: 505 LRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAY------- 557
++ ITI + G LS++E+ +MV++A + +D+E + + +NS + Y
Sbjct: 1572TGKEQQITIRSS-GGLSEDEIDKMVKEAELHAQKDQERKALIDIRNSADTSIYSIEKSLG 1748
Query: 558 EARDRV-KKLEKMVEEVIEWLDRNQLA--EAEEFEYKKQELEK 597
E RD++ ++ K +E+ + L R +A A+E + K K
Sbjct: 1749EYRDKIPSEVAKEIEDAVSDL-RTAMAGDNADEIKAKLDAANK 1874
Score = 27.7 bits (60), Expect(2) = e-131
Identities = 12/14 (85%), Positives = 13/14 (92%)
Frame = +1
Query: 10 IGIDLGTTYSCVAV 23
IGIDLGTT SCV+V
Sbjct: 49 IGIDLGTTNSCVSV 90
>TC214819 homologue to UP|Q8GSN2 (Q8GSN2) Cell-autonomous heat shock cognate
protein 70, partial (54%)
Length = 1266
Score = 372 bits (954), Expect = e-103
Identities = 196/315 (62%), Positives = 241/315 (76%), Gaps = 14/315 (4%)
Frame = +1
Query: 296 GIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQL 355
GID + TVT+ FE+LN DLF KCME VEKCL +AK+ K V ++VLVGGSTRIPKVQQL
Sbjct: 7 GIDFYSTVTRARFEELNMDLFRKCMEPVEKCLRDAKMDKRSVDDVVLVGGSTRIPKVQQL 186
Query: 356 LKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEV 415
L++ F+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLSLG+E
Sbjct: 187 LQDFFNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLET 354
Query: 416 VEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFS 475
GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+V+EGE A+T N LGKFELSG
Sbjct: 355 AGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIP 534
Query: 476 PAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQY 535
PAP+G +I VCFD+DA+GI+ VSAED + K ITITN GRLS+E++ +MV++A +Y
Sbjct: 535 PAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEKY 714
Query: 536 KAEDEEVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEWLDRNQ 581
K+EDEE +KKV+AKN+LENYAY R+ V KK+E +E+ I+WLD NQ
Sbjct: 715 KSEDEEHKKKVEAKNALENYAYNMRNTVKDDKIGEKLDPTDKKKIEDAIEQAIQWLDSNQ 894
Query: 582 LAEAEEFEYKKQELE 596
LAEA+EFE K +ELE
Sbjct: 895 LAEADEFEDKMKELE 939
>TC214820 homologue to UP|Q9M6R1 (Q9M6R1) High molecular weight heat shock
protein, partial (54%)
Length = 1272
Score = 296 bits (759), Expect = 1e-80
Identities = 158/260 (60%), Positives = 195/260 (74%), Gaps = 14/260 (5%)
Frame = +2
Query: 351 KVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLS 410
K QQL+++ ++ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLS
Sbjct: 164 K*QQLVQDFYNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLS 331
Query: 411 LGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFE 470
LG+E GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE A+T N LGKFE
Sbjct: 332 LGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTRDNNLLGKFE 511
Query: 471 LSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVR 530
LSG PAP+G +I VCFD+DA+GI+ VSAED + K ITITN GRLS++E+ +MV+
Sbjct: 512 LSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQ 691
Query: 531 DAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEW 576
+A +YKAEDEE +KKV AKN+LENYAY R+ + KK+E +E I+W
Sbjct: 692 EAEKYKAEDEEHKKKVDAKNALENYAYNMRNTIKDEKIASKLSDDDKKKIEDAIESAIQW 871
Query: 577 LDRNQLAEAEEFEYKKQELE 596
LD NQLAEA+EFE K +ELE
Sbjct: 872 LDGNQLAEADEFEDKMKELE 931
Score = 82.4 bits (202), Expect = 5e-16
Identities = 40/72 (55%), Positives = 51/72 (70%)
Frame = +1
Query: 297 IDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLL 356
+D + T+T+ FE+LN DLF KCME VEKCL +AK+ KS VH++VLVGGSTRIPKV +
Sbjct: 1 VDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVAAIG 180
Query: 357 KEMFSVHGRVKE 368
G V+E
Sbjct: 181 AGFLQWQGAVQE 216
>TC204156 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (40%)
Length = 965
Score = 249 bits (635), Expect = 3e-66
Identities = 129/218 (59%), Positives = 162/218 (74%), Gaps = 14/218 (6%)
Frame = +2
Query: 393 EGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKV 452
EG++KV++LLLLDV PLSLG+E GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+V
Sbjct: 2 EGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQV 181
Query: 453 YEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTIT 512
YEGE +T N LGKFELSG PAP+G +I VCFD+DA+GI+ VSAED + K IT
Sbjct: 182 YEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKIT 361
Query: 513 ITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRV--------- 563
ITN GRLS+EE+ +MV++A +YKAEDEE +KKV+AKN+LENYAY R+ +
Sbjct: 362 ITNDKGRLSKEEIEKMVQEAEKYKAEDEEHKKKVEAKNTLENYAYNMRNTIKDDKIASKL 541
Query: 564 -----KKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELE 596
KK+E +E+ I+WLD NQLAEA+EFE K +ELE
Sbjct: 542 SADDKKKIEDAIEQAIQWLDGNQLAEADEFEDKMKELE 655
>TC209139 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (45%)
Length = 733
Score = 227 bits (579), Expect = 1e-59
Identities = 116/165 (70%), Positives = 133/165 (80%)
Frame = +2
Query: 433 KKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDA 492
K+E T DNQTSV IKV+EGE AKT+ GKFELSGF+P+P+G +INV FDVD
Sbjct: 2 KRESXXSTFXDNQTSVLIKVFEGERAKTEDIXXXGKFELSGFTPSPRGVPQINVGFDVDV 181
Query: 493 DGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSL 552
DGIVEV+A D S LKK ITI+N+ GRLS EEMRRMVRDA +YKAEDEEVR KV+ KN L
Sbjct: 182 DGIVEVTARDRSTGLKKKITISNKHGRLSPEEMRRMVRDAVRYKAEDEEVRNKVRIKNLL 361
Query: 553 ENYAYEARDRVKKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELEK 597
ENYA+E RDRVK LEK+VEE IEWLDRNQLAE +EFEYK+QELE+
Sbjct: 362 ENYAFEMRDRVKNLEKVVEETIEWLDRNQLAETDEFEYKRQELEE 496
>TC204159 homologue to UP|Q40151 (Q40151) Hsc70 protein, partial (22%)
Length = 769
Score = 224 bits (570), Expect = 1e-58
Identities = 110/146 (75%), Positives = 123/146 (83%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 80 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 259
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ DMKLWPFKV+ G DKPMI V YKG+E++ +A
Sbjct: 260 QVAMNPINTVFDAKRLIGRRFSDASVQSDMKLWPFKVIPGPADKPMIVVNYKGDEKQFSA 439
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRA 146
EEISSMVL KMKE+AEAYLG +K A
Sbjct: 440 EEISSMVLIKMKEIAEAYLGSTIKNA 517
>TC205202 homologue to UP|HS7M_PEA (P37900) Heat shock 70 kDa protein,
mitochondrial precursor, partial (51%)
Length = 1228
Score = 215 bits (547), Expect = 5e-56
Identities = 129/307 (42%), Positives = 188/307 (61%), Gaps = 8/307 (2%)
Frame = +2
Query: 295 GGIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQ 354
G L++T+T+ FE L L E+ + CL +A I +V E++LVGG TR+PKVQ+
Sbjct: 20 GAKHLNITLTRSKFEALVNHLIERTKVPCKSCLKDANISIKDVDEVLLVGGMTRVPKVQE 199
Query: 355 LLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIE 414
++ E+F K K +NPDEAVA GAA+Q IL + V+ELLLLDV PLSLGIE
Sbjct: 200 VVSEIFG-----KSPSKGVNPDEAVAMGAAIQGGILRGD----VKELLLLDVTPLSLGIE 352
Query: 415 VVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGF 474
+ G+ + LI +NTTIPTKK + F T +DNQT V IKV +GE N LG+F+L G
Sbjct: 353 TLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKMLGEFDLVGI 532
Query: 475 SPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQ 534
PAP+G +I V FD+DA+GIV VSA+D S ++ ITI + G LS++E+ +MV++A
Sbjct: 533 PPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSS-GGLSEDEIEKMVKEAEL 709
Query: 535 YKAEDEEVRKKVKAKNSLENYAY-------EARDRV-KKLEKMVEEVIEWLDRNQLAEAE 586
+ +D+E + + +NS + Y E RD++ ++ K +E+ + L R ++E
Sbjct: 710 HAQKDQERKALIDIRNSADTTIYSIEKSLGEYRDKIPSEVAKEIEDAVSDL-RKAMSEDN 886
Query: 587 EFEYKKQ 593
E K +
Sbjct: 887 VDEIKSK 907
>TC234487 similar to UP|Q9SAU8 (Q9SAU8) HSP70, partial (23%)
Length = 457
Score = 211 bits (538), Expect = 6e-55
Identities = 105/152 (69%), Positives = 125/152 (82%)
Frame = +1
Query: 32 IPNELGNRTTPSYVAFTDTERLMGDAAINQLALNPHNTVFDAKRLIGRRFSDRSVQQDMK 91
I N+ GN TTPS VAFTD +RL+G+AA NQ A NP NTVFDAKRLIGR+FSD +Q+D
Sbjct: 1 IHNDQGNNTTPSCVAFTDQQRLIGEAAKNQAATNPENTVFDAKRLIGRKFSDPVIQKDKM 180
Query: 92 LWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVLFKMKEVAEAYLGHEVKRAVITVP 151
LWPFKVV G DKPMIS+ YKG+E+ L AEE+SSMVL KM+E+AEAYL V+ AV+TVP
Sbjct: 181 LWPFKVVAGINDKPMISLNYKGQEKHLLAEEVSSMVLIKMREIAEAYLETPVENAVVTVP 360
Query: 152 AYFNNAQRQSTKDAGEIAGFDVMRIINEPTAA 183
AYFN++QR++T DAG IAG +VMRIINEPTAA
Sbjct: 361 AYFNDSQRKATIDAGAIAGLNVMRIINEPTAA 456
>BQ296289 homologue to PIR|T45517|T45 heat shock protein 70 cytosolic
[imported] - spinach, partial (21%)
Length = 421
Score = 200 bits (509), Expect = 1e-51
Identities = 99/140 (70%), Positives = 117/140 (82%)
Frame = +1
Query: 161 STKDAGEIAGFDVMRIINEPTAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDE 220
+TKDAG IAG +VMRIINEPTAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+E
Sbjct: 1 ATKDAGVIAGLNVMRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEE 180
Query: 221 GIFKVNATVGDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRIL 280
GIF+V AT GDT+LGG DFDN +VN V F+RKH KDI N ++LRRLR ACE+AKR L
Sbjct: 181 GIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKHKKDINGNPRALRRLRTACERAKRTL 360
Query: 281 SSTSQTTIELDSLCGGIDLH 300
SST+QTTIE+DSL G+D +
Sbjct: 361 SSTAQTTIEIDSLYEGVDFY 420
>TC226591 homologue to PRF|1909352A.0|445605|1909352A heat shock protein
hsp70. {Pisum sativum;} , partial (45%)
Length = 1338
Score = 191 bits (485), Expect = 8e-49
Identities = 108/261 (41%), Positives = 159/261 (60%)
Frame = +3
Query: 327 LVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQ 386
L +AK+ ++ E++LVGGSTRIP VQ+L+K++ V ++NPDE VA GAAVQ
Sbjct: 3 LRDAKLSFKDLDEVILVGGSTRIPAVQELVKKLTGKDPNV-----TVNPDEVVALGAAVQ 167
Query: 387 AAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQT 446
A +L+ + V +++LLDV PLSLG+E + GVM+ +IP+NTT+PT K F T +D QT
Sbjct: 168 AGVLAGD----VSDIVLLDVTPLSLGLETLGGVMTKIIPRNTTLPTSKSEVFSTAADGQT 335
Query: 447 SVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLR 506
SV I V +GE N LG F L G PAP+G +I V FD+DA+GI+ V+A D
Sbjct: 336 SVEINVLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVKFDIDANGILSVTAIDKGTG 515
Query: 507 LKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRVKKL 566
K+ ITIT L +E+ RMV +A ++ ED+E R + KN ++ Y+ ++K+L
Sbjct: 516 KKQDITITG-ASTLPSDEVERMVNEAEKFSKEDKEKRDAIDTKNQADSVVYQTEKQLKEL 692
Query: 567 EKMVEEVIEWLDRNQLAEAEE 587
V ++ +L E ++
Sbjct: 693 GDKVPGPVKEKVEAKLGELKD 755
>BF324184 homologue to PIR|S53126|S53 dnaK-type molecular chaperone hsp70 -
rice (fragment), partial (18%)
Length = 398
Score = 189 bits (479), Expect = 4e-48
Identities = 91/123 (73%), Positives = 104/123 (83%)
Frame = -3
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYV FTDTERL+GDAA N
Sbjct: 369 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKN 190
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRRFSD SVQ D+KLWPFKV+ G +KPMI V+YKGE+++ AA
Sbjct: 189 QVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVLSGPAEKPMIQVSYKGEDKQFAA 10
Query: 121 EEI 123
EEI
Sbjct: 9 EEI 1
>TC222877 weakly similar to UP|Q94614 (Q94614) Heat shock 70kDa protein
(Fragment), partial (19%)
Length = 809
Score = 175 bits (443), Expect = 6e-44
Identities = 104/231 (45%), Positives = 144/231 (62%), Gaps = 15/231 (6%)
Frame = +1
Query: 381 YGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRT 440
YGAAVQAA+LS +G V L+LLD+ PLSLG+ V +MSV+IP+NTTIP ++ +T+ T
Sbjct: 1 YGAAVQAALLS-KGIVNVPNLVLLDITPLSLGVSVQGDLMSVVIPRNTTIPVRRTKTYVT 177
Query: 441 TSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSA 500
T DNQ++V I+VYEGE + N LG F LSG PAP+G + FD+D +GI+ VSA
Sbjct: 178 TEDNQSAVMIEVYEGERTRASDNNLLGFFTLSGIPPAPRGH-PLYETFDIDENGILSVSA 354
Query: 501 EDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEAR 560
E+ S K ITITN RLS +E++RM+++A YKAED++ +K KA N L+ Y Y+ +
Sbjct: 355 EEESTGNKNEITITNEKERLSTKEIKRMIQEAEYYKAEDKKFLRKAKAMNDLDYYVYKIK 534
Query: 561 DRVKKLE-------KMVEEVIEWL--------DRNQLAEAEEFEYKKQELE 596
+ +KK + K E V + D NQ + FE +ELE
Sbjct: 535 NALKKKDISSKLCSKEKENVSSAIARATDLLEDNNQQDDIVVFEDNLKELE 687
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,872,751
Number of Sequences: 63676
Number of extensions: 239887
Number of successful extensions: 1252
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 1191
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1198
length of query: 602
length of database: 12,639,632
effective HSP length: 103
effective length of query: 499
effective length of database: 6,081,004
effective search space: 3034420996
effective search space used: 3034420996
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0118c.15


