

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118c.11
(259 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
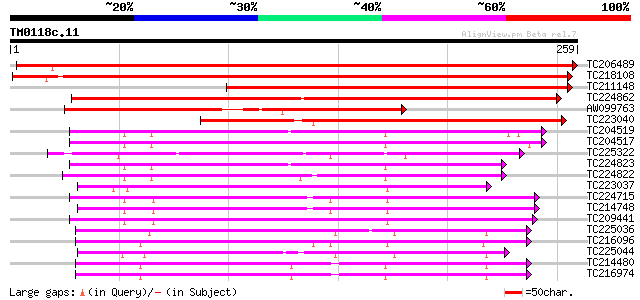
Score E
Sequences producing significant alignments: (bits) Value
TC206489 similar to UP|Q9XGG7 (Q9XGG7) Nodulin26-like major intr... 442 e-125
TC218108 UP|NO26_SOYBN (P08995) Nodulin-26 (N-26), complete 351 2e-97
TC211148 similar to UP|Q6QIL2 (Q6QIL2) NIP2, partial (58%) 259 8e-70
TC224862 similar to UP|Q6KAW0 (Q6KAW0) Nod26-like major intrinsi... 208 2e-54
AW099763 similar to PIR|JQ2285|JQ22 nodulin-26 - soybean, partia... 173 6e-44
TC223040 similar to UP|NI51_ARATH (Q9SV84) Probable aquaporin NI... 157 3e-39
TC204519 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic... 119 1e-27
TC204517 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic... 115 2e-26
TC225322 similar to UP|TI41_ARATH (O82316) Probable aquaporin TI... 113 1e-25
TC224823 similar to UP|TIP1_TOBAC (P21653) Probable aquaporin TI... 112 1e-25
TC224822 similar to UP|TIP2_TOBAC (P24422) Probable aquaporin TI... 112 1e-25
TC223037 similar to UP|Q8H1Z5 (Q8H1Z5) Tonoplast intrinsic prote... 103 1e-22
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 102 2e-22
TC214748 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 100 5e-22
TC209441 UP|Q39883 (Q39883) Nodulin-26, partial (98%) 100 1e-21
TC225036 similar to UP|O65357 (O65357) Aquaporin 2, partial (92%) 97 7e-21
TC216096 homologue to UP|PI27_ARATH (P93004) Aquaporin PIP2.7 (P... 96 2e-20
TC225044 homologue to UP|O22339 (O22339) Aquaporin-like transmem... 95 3e-20
TC214480 homologue to UP|Q39822 (Q39822) Pip1 protein, complete 94 6e-20
TC216974 homologue to UP|Q39822 (Q39822) Pip1 protein, complete 94 6e-20
>TC206489 similar to UP|Q9XGG7 (Q9XGG7) Nodulin26-like major intrinsic
protein, partial (86%)
Length = 1169
Score = 442 bits (1136), Expect = e-125
Identities = 216/257 (84%), Positives = 244/257 (94%), Gaps = 1/257 (0%)
Frame = +1
Query: 4 EVVLNVNNEASKKCD-SIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLP 62
+VVLNVN +A KKCD S +DCVPLLQKLVAEV+GTYFLIFAGCASVVVNL+ DKVVT P
Sbjct: 136 QVVLNVNGDAPKKCDDSANQDCVPLLQKLVAEVVGTYFLIFAGCASVVVNLDKDKVVTQP 315
Query: 63 GIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASG 122
GI+IVWGL VMVLVYS+GHISGAHFNPAVTIAH TTKRFPLKQVPAY++AQV+G+TLASG
Sbjct: 316 GISIVWGLTVMVLVYSVGHISGAHFNPAVTIAHATTKRFPLKQVPAYVIAQVVGATLASG 495
Query: 123 TLRLIFSGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGS 182
TLRLIF+GK++HF GTLP+GSDLQ+FVVEFIITF LMFV+SGVATDNRAIGELAGLAVGS
Sbjct: 496 TLRLIFNGKNDHFAGTLPSGSDLQSFVVEFIITFYLMFVISGVATDNRAIGELAGLAVGS 675
Query: 183 TVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITN 242
TV+LNV+FAGPITGASMNPARSLGPAIVH++Y+GIWIY+VSP LGAVAGTWAY+FIR TN
Sbjct: 676 TVLLNVMFAGPITGASMNPARSLGPAIVHHEYRGIWIYLVSPTLGAVAGTWAYNFIRYTN 855
Query: 243 KPVRELTKSSSFLKGAK 259
KPVRE+TKS+SFLKG++
Sbjct: 856 KPVREITKSASFLKGSE 906
>TC218108 UP|NO26_SOYBN (P08995) Nodulin-26 (N-26), complete
Length = 1122
Score = 351 bits (900), Expect = 2e-97
Identities = 168/259 (64%), Positives = 214/259 (81%), Gaps = 3/259 (1%)
Frame = +3
Query: 2 ADEVVLNVNNEASK---KCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV 58
+ EVV+NV S+ + DS+ VP LQKLVAE +GTYFLIFAGCAS+VVN N +
Sbjct: 120 SQEVVVNVTKNTSETIQRSDSLVS--VPFLQKLVAEAVGTYFLIFAGCASLVVNENYYNM 293
Query: 59 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGST 118
+T PGIAIVWGL + VLVY++GHISG HFNPAVTIA +T+RFPL QVPAY++AQ++GS
Sbjct: 294 ITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQVPAYVVAQLLGSI 473
Query: 119 LASGTLRLIFSGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGL 178
LASGTLRL+F G + F+GT+P G++LQAFV EFI+TF LMFV+ GVATDNRA+GELAG+
Sbjct: 474 LASGTLRLLFMGNHDQFSGTVPNGTNLQAFVFEFIMTFFLMFVICGVATDNRAVGELAGI 653
Query: 179 AVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFI 238
A+GST++LNV+ GP+TGASMNPARSLGPA VH +Y+GIWIY+++P++GA+AG W Y+ +
Sbjct: 654 AIGSTLLLNVIIGGPVTGASMNPARSLGPAFVHGEYEGIWIYLLAPVVGAIAGAWVYNIV 833
Query: 239 RITNKPVRELTKSSSFLKG 257
R T+KP+ E TKS+SFLKG
Sbjct: 834 RYTDKPLSETTKSASFLKG 890
>TC211148 similar to UP|Q6QIL2 (Q6QIL2) NIP2, partial (58%)
Length = 587
Score = 259 bits (662), Expect = 8e-70
Identities = 122/158 (77%), Positives = 148/158 (93%)
Frame = +2
Query: 100 RFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDLQAFVVEFIITFLLM 159
RFPLKQVP Y++AQV+GSTLAS TLRL+FSGK+ F+GTLP+GS+LQAFV+EF+ITF LM
Sbjct: 11 RFPLKQVPVYVVAQVVGSTLASATLRLLFSGKETQFSGTLPSGSNLQAFVIEFLITFFLM 190
Query: 160 FVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWI 219
FV+SGVATD+RAIGELA +AVGSTV+LNV+FAGPITGASMNPAR++GPAI+HN+Y+GIWI
Sbjct: 191 FVISGVATDDRAIGELAVIAVGSTVLLNVMFAGPITGASMNPARNIGPAILHNEYRGIWI 370
Query: 220 YMVSPILGAVAGTWAYSFIRITNKPVRELTKSSSFLKG 257
Y+VSP LGAVAGTW Y+ IR T+KP+RE+TKS+SFLKG
Sbjct: 371 YIVSPTLGAVAGTWVYNTIRYTDKPLREITKSTSFLKG 484
>TC224862 similar to UP|Q6KAW0 (Q6KAW0) Nod26-like major intrinsic protein,
partial (79%)
Length = 1192
Score = 208 bits (529), Expect = 2e-54
Identities = 103/224 (45%), Positives = 150/224 (65%)
Frame = +1
Query: 29 QKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+K+ AEV+GT+ L+F G S ++ ++ +V+ G ++ GL V V++YSIGHISGAH N
Sbjct: 274 RKVFAEVIGTFLLVFVGSGSAGLSKIDESMVSKLGASLAGGLIVTVMIYSIGHISGAHMN 453
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDLQAF 148
PAV++A + P Q+P Y+ AQ+ G+ AS TLR + D GT PAGS +QA
Sbjct: 454 PAVSLAFTAVRHLPWPQLPFYVAAQLTGAISASYTLRELLRPSDE-IGGTSPAGSHIQAL 630
Query: 149 VVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPA 208
++E + T+ ++F+ VATD+ A G+L+G+AVGS+V + + AGPI+G SMNPAR+LGPA
Sbjct: 631 IMEMVSTYTMVFISMAVATDSNATGQLSGVAVGSSVCIASIVAGPISGGSMNPARTLGPA 810
Query: 209 IVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTKSS 252
I + YKG+W+Y V PI GAV W+Y+ IR T P ++ SS
Sbjct: 811 IATSYYKGLWVYFVGPITGAVLAAWSYNVIRDTEHPGFPISLSS 942
>AW099763 similar to PIR|JQ2285|JQ22 nodulin-26 - soybean, partial (34%)
Length = 472
Score = 173 bits (439), Expect = 6e-44
Identities = 98/167 (58%), Positives = 118/167 (69%), Gaps = 11/167 (6%)
Frame = +2
Query: 26 PLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGA 85
PLL KLVAEV+GTYFLIFAGCASVV NL+ DKVVT PGI+IVWGL VMVLVYS GHISGA
Sbjct: 2 PLLHKLVAEVVGTYFLIFAGCASVVENLDKDKVVTQPGISIVWGLTVMVLVYSFGHISGA 181
Query: 86 HFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGT-----------LRLIFSGKDNH 134
HF PAVTIAH + P + + G++L + L L GK++
Sbjct: 182 HFYPAVTIAHSS---------PP*VFTET-GTSLCDNSGSSEPH*LVELLDLYSIGKNDS 331
Query: 135 FTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVG 181
F GTLP+ SD+Q+FVVE+IIT LM+V++GVATD+ +IGELA LA G
Sbjct: 332 FAGTLPSCSDVQSFVVEYIITIYLMYVIAGVATDHISIGELAWLAAG 472
>TC223040 similar to UP|NI51_ARATH (Q9SV84) Probable aquaporin NIP5.1
(NOD26-like intrinsic protein 5.1) (Nodulin-26-like
major intrinsic protein 6) (AtNLM6) (NLM6 protein)
(NodLikeMip6), partial (56%)
Length = 869
Score = 157 bits (398), Expect = 3e-39
Identities = 82/169 (48%), Positives = 112/169 (65%), Gaps = 2/169 (1%)
Frame = +1
Query: 88 NPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTG--TLPAGSDL 145
NP++TIA + FP VPAYI AQV S A L+ ++ +G T+P S
Sbjct: 10 NPSLTIAFAAFRHFPWTHVPAYIAAQVSASICACYALKGVYHP---FLSGGVTVPTVSVA 180
Query: 146 QAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSL 205
QAF EFIITF+L+FVV+ VATD RA+GELAG+AVG+TV+LN+L +GP +G SMNP R+L
Sbjct: 181 QAFATEFIITFILLFVVTAVATDTRAVGELAGIAVGATVLLNILISGPTSGGSMNPVRTL 360
Query: 206 GPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTKSSSF 254
GPA+ YK IWIY+V+P LGA+AG Y+ +++ ++ + SF
Sbjct: 361 GPAVAAGNYKHIWIYLVAPTLGALAGAGVYTLVKLRDEEAEPPRQVRSF 507
>TC204519 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic protein,
partial (97%)
Length = 989
Score = 119 bits (299), Expect = 1e-27
Identities = 79/227 (34%), Positives = 118/227 (51%), Gaps = 9/227 (3%)
Frame = +1
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 82
++ +AE + T +FAG S + L +D + G +AI G A+ V V +I
Sbjct: 112 IKAYIAEFISTLLFVFAGVGSAIAYAKLTSDAALDPTGLVAVAICHGFALFVAVSVGANI 291
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT + Y +AQ++GS +AS L+ + +G D
Sbjct: 292 SGGHVNPAVTFGLALGGHITILTGLFYWIAQLLGSIVASLLLKFV-TGYDTPIHSVAAGI 468
Query: 143 SDLQAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITGASMN 200
+ V E IITF L++ V A D + ++G +A +A+G V N+L AGP +G SMN
Sbjct: 469 GAGEGVVTEIIITFGLVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGGSMN 648
Query: 201 PARSLGPAIVHNQYKGIWIYMVSPIL-GAVAG-TWAYSFIRITNKPV 245
PARS GPA+V + WIY V P++ G +AG + Y+FI + P+
Sbjct: 649 PARSFGPAVVSGDFHDNWIYWVGPLIGGGLAGLIYTYAFIPTNHAPL 789
>TC204517 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic protein,
complete
Length = 1016
Score = 115 bits (288), Expect = 2e-26
Identities = 76/227 (33%), Positives = 110/227 (47%), Gaps = 9/227 (3%)
Frame = +3
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 82
++ +AE T +FAG S + L +D + G +AI G A+ V V +I
Sbjct: 120 IKAYIAEFHSTLLFVFAGVGSAIAYGKLTSDAALDPAGLLAVAICHGFALFVAVSVGANI 299
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT + Y +AQ++GS +A L + G
Sbjct: 300 SGGHVNPAVTFGLALGGHITILTGFFYWIAQLLGSIVACFLLNYVTGGLPTPIHSVASGV 479
Query: 143 SDLQAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITGASMN 200
++ V E IITF L++ V A D + ++G +A +A+G V N+L AGP +G SMN
Sbjct: 480 GAVEGVVTEIIITFGLVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGGSMN 659
Query: 201 PARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYS--FIRITNKPV 245
PARS GPA+V + WIY V P++G Y FIR + P+
Sbjct: 660 PARSFGPAVVSGDFHDNWIYWVGPLIGGGLAGLIYGNVFIRSDHAPL 800
>TC225322 similar to UP|TI41_ARATH (O82316) Probable aquaporin TIP4.1
(Tonoplast intrinsic protein 4.1) (Epsilon-tonoplast
intrinsic protein) (Epsilon-TIP), partial (98%)
Length = 1378
Score = 113 bits (282), Expect = 1e-25
Identities = 77/224 (34%), Positives = 113/224 (50%), Gaps = 6/224 (2%)
Frame = +2
Query: 18 DSIEEDCVPLLQKLVAEVLGTYFLIFAGCAS--VVVNLNNDKVVTLPGIAIVWGLAVMVL 75
++ + DC+ Q L+ E + T+ +F G S VV L D +V L +A+ L V V+
Sbjct: 575 EATQPDCI---QALIVEFIATFLFVFVGVGSSMVVDKLGGDALVGLFAVAVAHALVVAVM 745
Query: 76 VYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHF 135
+ S HISG H NPAVT+ + + + Y + Q++ + AS L + G+
Sbjct: 746 I-SAAHISGGHLNPAVTLGLLAGGHITIFRSLLYWIDQLVAAAAASYLLYYLSGGQATP- 919
Query: 136 TGTLPAGSDL-QAFVVEFIITFLLMFVVSGVATDNRAIGELAGLA---VGSTVMLNVLFA 191
TL +G Q V E ++TF L+F V D + G LAGL VG V N+L
Sbjct: 920 VHTLASGVGYGQGVVWEIVLTFSLLFTVYATMVDPKK-GALAGLGPTLVGFVVGANILAG 1096
Query: 192 GPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAY 235
G + ASMNPARS GPA+V + W+Y V P++G + Y
Sbjct: 1097GAYSAASMNPARSFGPALVTGNWTDHWVYWVGPLIGGGLAGFIY 1228
>TC224823 similar to UP|TIP1_TOBAC (P21653) Probable aquaporin TIP-type
RB7-5A (Tonoplast intrinsic protein, root-specific
RB7-5A) (TobRB7) (RT-TIP), partial (96%)
Length = 1083
Score = 112 bits (281), Expect = 1e-25
Identities = 76/207 (36%), Positives = 104/207 (49%), Gaps = 7/207 (3%)
Frame = +2
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 82
L+ AE T +FAG S + L D + G +A+ A+ V V +I
Sbjct: 86 LKAYFAEFHATLIFVFAGVGSAIAYNELTKDAALDPTGLVAVAVAHAFALFVGVSVAANI 265
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT L Y +AQ++GS +A L LI + K
Sbjct: 266 SGGHLNPAVTFGLAIGGNITLITGFLYWIAQLLGSIVACLLLNLI-TAKSIPSHSPANGV 442
Query: 143 SDLQAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITGASMN 200
+DLQA V E +ITF L++ V A D + ++G +A +A+G V N+L AGP +G SMN
Sbjct: 443 NDLQAVVFEIVITFGLVYTVYATAVDPKKGSLGIIAPIAIGFVVGANILAAGPFSGGSMN 622
Query: 201 PARSLGPAIVHNQYKGIWIYMVSPILG 227
PARS GPA+V WIY V P++G
Sbjct: 623 PARSFGPAVVSGDLAANWIYWVGPLIG 703
>TC224822 similar to UP|TIP2_TOBAC (P24422) Probable aquaporin TIP-type
RB7-18C (Tonoplast intrinsic protein, root-specific
RB7-18C) (TobRB7) (RT-TIP), partial (96%)
Length = 1202
Score = 112 bits (281), Expect = 1e-25
Identities = 76/211 (36%), Positives = 107/211 (50%), Gaps = 8/211 (3%)
Frame = +1
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSI 79
V L+ +AE T +FAG S + L D + G +A+ A+ V V
Sbjct: 142 VASLKAYLAEFHATLIFVFAGVGSAIAYNELTKDAALDPTGLVAVAVAHAFALFVGVSVA 321
Query: 80 GHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK-DNHFTGT 138
+ISG H NPAVT L Y +AQ++GS +A L I + +H T
Sbjct: 322 ANISGGHLNPAVTFGLAIGGNITLITGFLYWIAQLLGSIVACLLLNFITAKSIPSHAPAT 501
Query: 139 LPAGSDLQAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITG 196
+D QA V E +ITF L++ V A D + ++G +A +A+G V N+L AGP +G
Sbjct: 502 --GVNDFQAVVFEIVITFGLVYTVYATAADPKKGSLGIIAPIAIGFVVGANILAAGPFSG 675
Query: 197 ASMNPARSLGPAIVHNQYKGIWIYMVSPILG 227
SMNPARS GPA+V + WIY V P++G
Sbjct: 676 GSMNPARSFGPAVVSGDFAANWIYWVGPLIG 768
>TC223037 similar to UP|Q8H1Z5 (Q8H1Z5) Tonoplast intrinsic protein
bobTIP26-2, partial (76%)
Length = 723
Score = 103 bits (256), Expect = 1e-22
Identities = 69/195 (35%), Positives = 98/195 (49%), Gaps = 6/195 (3%)
Frame = +3
Query: 32 VAEVLGTYFLIFAGC-ASVVVN-LNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNP 89
++E + T +FAG +SV VN L DK L A+ A+ V V +ISG H NP
Sbjct: 138 LSEFISTLIFVFAGSGSSVAVNKLTVDKPSALVVAAVAHAFALFVAVSVSTNISGGHVNP 317
Query: 90 AVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDLQAFV 149
AVT L + + +AQ++GS +A L+ I G+D A V
Sbjct: 318 AVTFGAFVGGNLTLLRCVLFWIAQILGSVIACLLLKFITGGQDVPVFKLSSGVGVGNAVV 497
Query: 150 VEFIITFLLMFVVSGVATDNRA----IGELAGLAVGSTVMLNVLFAGPITGASMNPARSL 205
+E ++TF L++ V D R+ +G +A + +G V NVL GP GASMNPA S
Sbjct: 498 LEMVMTFGLVYTVYATTVDPRSRRGSLGVMAPIVIGFIVGANVLVGGPFDGASMNPAASF 677
Query: 206 GPAIVHNQYKGIWIY 220
GPA+V +K W+Y
Sbjct: 678 GPAVVGWSWKNHWVY 722
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 102 bits (253), Expect = 2e-22
Identities = 70/223 (31%), Positives = 112/223 (49%), Gaps = 8/223 (3%)
Frame = +1
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPGI---AIVWGLAVMVLVYSIGHI 82
L+ +AE + T+ +FAG S + L ++ T G+ +I A+ V V +I
Sbjct: 559 LKAALAEFISTFIFVFAGSGSGIAYNKLTDNGAATPAGLISASIAHAFALFVAVSVGANI 738
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT + Y++AQ++GS +AS L + + F L AG
Sbjct: 739 SGGHVNPAVTFGAFVGGNITFLRGIVYVIAQLLGSIVASLLLAFVTASTVPAFG--LSAG 912
Query: 143 SDL-QAFVVEFIITFLLMFVVSGVATDNRA--IGELAGLAVGSTVMLNVLFAGPITGASM 199
+ A V+E ++TF L++ V A D + +G +A +A+G V N+L G +GA+M
Sbjct: 913 VGVGNALVLEIVMTFGLVYTVYATAIDPKKGNLGIIAPIAIGFIVGANILLGGAFSGAAM 1092
Query: 200 NPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITN 242
NPA + GPA+V + WIY P++G Y + I++
Sbjct: 1093NPAVTFGPAVVSWTWTNHWIYWAGPLIGGGIAGLVYEVVFISH 1221
>TC214748 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1254
Score = 100 bits (250), Expect = 5e-22
Identities = 70/219 (31%), Positives = 110/219 (49%), Gaps = 8/219 (3%)
Frame = +3
Query: 32 VAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPGI---AIVWGLAVMVLVYSIGHISGAH 86
+AE + T +FAG S + L ++ T G+ +I A+ V V +ISG H
Sbjct: 198 LAEFISTLIFVFAGSGSGIAYNKLTDNGAATPAGLISASIAHAFALFVAVSVGANISGGH 377
Query: 87 FNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDL- 145
NPAVT L + Y++AQ++GS +AS L + + F L AG +
Sbjct: 378 VNPAVTFGAFVGGNITLLRGIVYVIAQLLGSIVASLLLAFVTASPVPAFG--LSAGVGVG 551
Query: 146 QAFVVEFIITFLLMFVVSGVATDNRA--IGELAGLAVGSTVMLNVLFAGPITGASMNPAR 203
A V+E ++TF L++ V A D + +G +A +A+G V N+L G +GA+MNPA
Sbjct: 552 NALVLEIVMTFGLVYTVYATAVDPKKGNLGIIAPIAIGFIVGANILLGGAFSGAAMNPAV 731
Query: 204 SLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITN 242
+ GPA+V + WIY P++G Y + I++
Sbjct: 732 TFGPAVVSWTWTNHWIYWAGPLIGGGIAGLIYEVVFISH 848
>TC209441 UP|Q39883 (Q39883) Nodulin-26, partial (98%)
Length = 982
Score = 99.8 bits (247), Expect = 1e-21
Identities = 70/221 (31%), Positives = 102/221 (45%), Gaps = 7/221 (3%)
Frame = +2
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPGI---AIVWGLAVMVLVYSIGHI 82
L+ +AE + +FAG S + L N+ T G+ ++ A+ V V +I
Sbjct: 131 LKAALAEFISMLIFVFAGEGSGMAYNKLTNNGSATPAGLVAASLSHAFALFVAVSVGANI 310
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT L + Y +AQ++GS +A L+ G + P
Sbjct: 311 SGGHVNPAVTFGAFVGGHITLFRSILYWIAQLLGSVVACLLLKFATGGLETSAFALSPGV 490
Query: 143 SDLQAFVVEFIITFLLMFVVSGVATDNRA--IGELAGLAVGSTVMLNVLFAGPITGASMN 200
A V E ++TF L++ V A D + +G +A +A+G V N+L G GASMN
Sbjct: 491 EAGNALVFEIVMTFGLVYTVYATAVDPKKGDLGIIAPIAIGFIVGANILAGGAFDGASMN 670
Query: 201 PARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRIT 241
PA S GPA+V + W+Y V P GA Y I+
Sbjct: 671 PAVSFGPAVVSWTWSNHWVYWVGPFAGAAIAAIVYEIFFIS 793
>TC225036 similar to UP|O65357 (O65357) Aquaporin 2, partial (92%)
Length = 1610
Score = 97.1 bits (240), Expect = 7e-21
Identities = 69/228 (30%), Positives = 107/228 (46%), Gaps = 20/228 (8%)
Frame = +3
Query: 31 LVAEVLGTYFLIFAGCASVVVNLNNDKVVTLP-------GIAIVWGLAVMVLVYSIGHIS 83
L+AE + T ++ +V+ + P GIA +G + VLVY IS
Sbjct: 597 LIAEFVATLLFLYVTILTVIGYNHQTATAAEPCSGVGVLGIAWAFGGMIFVLVYCTAGIS 776
Query: 84 GAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGS 143
G H NPAVT ++ L + Y++AQV+G+ G ++ + N + G + +
Sbjct: 777 GGHINPAVTFGLFLARKVSLTRAVGYMVAQVLGAISGVGLVKALQKSYYNRYKGGVNMLA 956
Query: 144 DLQA----FVVEFIITFLLMFVVSGVATDNRAIGE------LAGLAVGSTVMLNVLFAGP 193
D + E I TF+L++ V ATD + + LA L +G V + L P
Sbjct: 957 DGYSKGTGLGAEIIGTFILVYTVFS-ATDPKRVARDSHVPVLAPLPIGFAVFMVHLATIP 1133
Query: 194 ITGASMNPARSLGPAIVHNQYKG---IWIYMVSPILGAVAGTWAYSFI 238
ITG +NPARSLGPA++ N K WI+ V P +GA + + +
Sbjct: 1134ITGTGINPARSLGPAVIFNNEKAWDDQWIFWVGPFIGAALAAFYHQSV 1277
>TC216096 homologue to UP|PI27_ARATH (P93004) Aquaporin PIP2.7 (Plasma
membrane intrinsic protein 3) (Salt-stress induced major
intrinsis protein), partial (97%)
Length = 1043
Score = 95.9 bits (237), Expect = 2e-20
Identities = 71/223 (31%), Positives = 107/223 (47%), Gaps = 15/223 (6%)
Frame = +1
Query: 31 LVAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHF 87
L+AE + T ++ A+V+ + V L GIA +G + VLVY ISG H
Sbjct: 100 LIAEFIATLLFLYVTVATVIGHKKQTGPCDGVGLLGIAWAFGGMIFVLVYCTAGISGGHI 279
Query: 88 NPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTG---TLPAGSD 144
NPAVT ++ L + Y++AQ +G+ G ++ N G ++ AG +
Sbjct: 280 NPAVTFGLFLARKVSLIRALFYMVAQCLGAICGVGLVKAFMKHSYNSLGGGANSVSAGYN 459
Query: 145 L-QAFVVEFIITFLLMFVVSGVATDNRA-----IGELAGLAVGSTVMLNVLFAGPITGAS 198
A E I TF+L++ V R+ I LA L +G V + L PITG
Sbjct: 460 KGSALGAEIIGTFVLVYTVFSATDPKRSARDSHIPVLAPLPIGFAVFMVHLATIPITGTG 639
Query: 199 MNPARSLGPAIVHNQYK---GIWIYMVSPILGAVAGTWAYSFI 238
+NPARS G A+++N K WI+ V P +GA+A + +I
Sbjct: 640 INPARSFGAAVIYNNGKVWDDHWIFWVGPFVGALAAAAYHQYI 768
>TC225044 homologue to UP|O22339 (O22339) Aquaporin-like transmembrane
channel protein, complete
Length = 1235
Score = 95.1 bits (235), Expect = 3e-20
Identities = 71/216 (32%), Positives = 108/216 (49%), Gaps = 19/216 (8%)
Frame = +2
Query: 32 VAEVLGTYFLIFAGCASVV-VNLNNDKVVT--LPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+AE + T+ ++ +V+ VN ++ K T + GIA +G + LVY ISG H N
Sbjct: 203 IAEFVATFLFLYITILTVMGVNRSSSKCATVGIQGIAWAFGGMIFALVYCTAGISGGHIN 382
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDLQA- 147
PAVT ++ L + Y++ QV+G+ + +G ++ F GK F G G++ A
Sbjct: 383 PAVTFGLFLARKLSLTRALFYMVMQVLGAIVGAGVVK-GFEGKT--FYGQHNGGANFVAP 553
Query: 148 -------FVVEFIITFLLMFVVSGVATDNRAIGE-----LAGLAVGSTVMLNVLFAGPIT 195
E + TF+L++ V R+ + LA L +G V L L PIT
Sbjct: 554 GYTKGDGLGAEIVGTFILVYTVFSATDAKRSARDSHVPILAPLPIGFAVFLVHLATIPIT 733
Query: 196 GASMNPARSLGPAIVHNQYKG---IWIYMVSPILGA 228
G +NPARSLG AI+ N+ G WI+ V P +GA
Sbjct: 734 GTGINPARSLGAAIIFNKDLGWDDHWIFWVGPFVGA 841
Score = 35.0 bits (79), Expect = 0.034
Identities = 28/102 (27%), Positives = 46/102 (44%), Gaps = 9/102 (8%)
Frame = +2
Query: 139 LPAGSDLQAFVVEFIITFLLMFV-VSGVATDNRAIGELAGL-------AVGSTVMLNVLF 190
L + S +A + EF+ TFL +++ + V NR+ + A + A G + V
Sbjct: 173 LTSWSFYRAGIAEFVATFLFLYITILTVMGVNRSSSKCATVGIQGIAWAFGGMIFALVYC 352
Query: 191 AGPITGASMNPARSLGPAIVHN-QYKGIWIYMVSPILGAVAG 231
I+G +NPA + G + YMV +LGA+ G
Sbjct: 353 TAGISGGHINPAVTFGLFLARKLSLTRALFYMVMQVLGAIVG 478
>TC214480 homologue to UP|Q39822 (Q39822) Pip1 protein, complete
Length = 1240
Score = 94.0 bits (232), Expect = 6e-20
Identities = 69/232 (29%), Positives = 106/232 (44%), Gaps = 24/232 (10%)
Frame = +1
Query: 31 LVAEVLGTYFLIFAGCASVVVNLNNDKV---------VTLPGIAIVWGLAVMVLVYSIGH 81
L+AE + T ++ +V+ + V V + GIA +G + +LVY
Sbjct: 214 LIAEFIATMLFLYITVLTVIGYKSQSDVKAGGDVCGGVGILGIAWAFGGMIFILVYCTAG 393
Query: 82 ISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLI-------FSGKDNH 134
ISG H NPAVT ++ L + Y++AQ +G+ G ++ + G N
Sbjct: 394 ISGGHINPAVTFGLFLARKVSLIRAIMYMVAQCLGAICGVGLVKAFQKAYYNRYGGGANE 573
Query: 135 FTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNR-----AIGELAGLAVGSTVMLNVL 189
+ G L A E I TF+L++ V R + LA L +G V + L
Sbjct: 574 LSEGYSTGVGLGA---EIIGTFVLVYTVFSATDPKRNARDSHVPVLAPLPIGFAVFMVHL 744
Query: 190 FAGPITGASMNPARSLGPAIVHNQYKG---IWIYMVSPILGAVAGTWAYSFI 238
P+TG +NPARSLG A+++NQ K WI+ V P +GA + + FI
Sbjct: 745 ATIPVTGTGINPARSLGAAVMYNQQKAWDDHWIFWVGPFIGAAIAAFYHQFI 900
>TC216974 homologue to UP|Q39822 (Q39822) Pip1 protein, complete
Length = 1254
Score = 94.0 bits (232), Expect = 6e-20
Identities = 69/232 (29%), Positives = 106/232 (44%), Gaps = 24/232 (10%)
Frame = +3
Query: 31 LVAEVLGTYFLIFAGCASVVVNLNNDKV---------VTLPGIAIVWGLAVMVLVYSIGH 81
L+AE + T ++ +V+ + V V + GIA +G + +LVY
Sbjct: 210 LIAEFIATMLFLYITVLTVIGYKSQSDVKAGGDVCGGVGILGIAWAFGGMIFILVYCTAG 389
Query: 82 ISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLI-------FSGKDNH 134
ISG H NPAVT ++ L + Y++AQ +G+ G ++ + G N
Sbjct: 390 ISGGHINPAVTFGLFLARKVSLIRAIMYMVAQCLGAICGVGLVKAFQKAYYNRYGGGANE 569
Query: 135 FTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNR-----AIGELAGLAVGSTVMLNVL 189
+ G L A E I TF+L++ V R + LA L +G V + L
Sbjct: 570 LSEGYSTGVGLGA---EIIGTFVLVYTVFSATDPKRNARDSHVPVLAPLPIGFAVFMVHL 740
Query: 190 FAGPITGASMNPARSLGPAIVHNQYKG---IWIYMVSPILGAVAGTWAYSFI 238
P+TG +NPARSLG A+++NQ K WI+ V P +GA + + FI
Sbjct: 741 ATIPVTGTGINPARSLGAAVMYNQQKAWDDHWIFWVGPFIGAAIAAFYHQFI 896
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,922,947
Number of Sequences: 63676
Number of extensions: 121379
Number of successful extensions: 681
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 625
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 644
length of query: 259
length of database: 12,639,632
effective HSP length: 95
effective length of query: 164
effective length of database: 6,590,412
effective search space: 1080827568
effective search space used: 1080827568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0118c.11


