

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
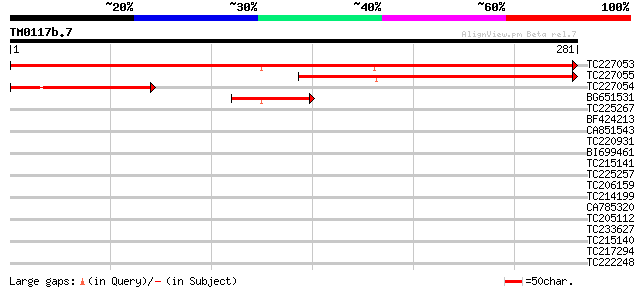
Query= TM0117b.7
(281 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227053 similar to GB|AAP21363.1|30102890|BT006555 At4g38290 {A... 438 e-123
TC227055 similar to GB|AAP21363.1|30102890|BT006555 At4g38290 {A... 225 2e-59
TC227054 119 2e-27
BG651531 66 2e-11
TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucos... 32 0.42
BF424213 weakly similar to GP|21322711|em pherophorin-dz1 protei... 31 0.72
CA851543 31 0.72
TC220931 similar to UP|Q71QD5 (Q71QD5) Protein phosphatase 2C, p... 30 0.94
BI699461 30 1.2
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 30 1.2
TC225257 similar to UP|Q7XXS4 (Q7XXS4) Thiamine biosynthetic enz... 30 1.6
TC206159 similar to UP|SY71_ARATH (Q9SF29) Syntaxin 71 (AtSYP71)... 30 1.6
TC214199 weakly similar to UP|Q6NLH4 (Q6NLH4) At5g54470, partial... 29 2.1
CA785320 29 2.1
TC205112 similar to UP|Q93ZP1 (Q93ZP1) AT5g61020/maf19_20, parti... 29 2.7
TC233627 weakly similar to UP|Q7PKR5 (Q7PKR5) ENSANGP00000023757... 28 3.6
TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 28 3.6
TC217294 similar to UP|Q8SKU2 (Q8SKU2) Tic62 protein precursor, ... 28 3.6
TC222248 weakly similar to GB|AAC51925.1|2708305|AF016369 U4/U6 ... 28 4.6
>TC227053 similar to GB|AAP21363.1|30102890|BT006555 At4g38290 {Arabidopsis
thaliana;} , partial (42%)
Length = 1356
Score = 438 bits (1126), Expect = e-123
Identities = 226/288 (78%), Positives = 245/288 (84%), Gaps = 7/288 (2%)
Frame = +1
Query: 1 MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS 60
MDLPNRDKQ TV VKKTA RDLQNDNKIM TSVGSSS KDKD TDS+R+SGTKRPLS
Sbjct: 214 MDLPNRDKQLTVAVKKTALRDLQNDNKIMVPTSVGSSSFFKDKDLGTDSSRVSGTKRPLS 393
Query: 61 DYPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETA 120
DYP+NHHLQQSP N+ ANGHLVYVRRKSEAEL K A+ENP+ NA+C RQLC EEETA
Sbjct: 394 DYPLNHHLQQSPSNNAANGHLVYVRRKSEAELSKGTAYENPSINAYCSHSRQLCCEEETA 573
Query: 121 LPK------PQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVT 174
PK PQLKEP SCFPAFAPFP+ SS++SSG+PSVPIS GKS KLAPVESN VT
Sbjct: 574 QPKSQIKEPPQLKEPKASCFPAFAPFPMASSMNSSGKPSVPISLGKSAKKLAPVESNYVT 753
Query: 175 TSSGP-TIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVEL 233
SSGP TIGNPKGL ++ W+ERYQQLQM LRKLDQSD+ EY+QML SLSSVELS+HAVEL
Sbjct: 754 ASSGPTTIGNPKGLKNLHWDERYQQLQMFLRKLDQSDREEYIQMLHSLSSVELSKHAVEL 933
Query: 234 EKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPADHDECSDKLKTSS 281
EKR+IQLSLEEAKELQRVAVLNVLGKS K FKAPAD+DECSDKLKT S
Sbjct: 934 EKRSIQLSLEEAKELQRVAVLNVLGKSVKNFKAPADYDECSDKLKTLS 1077
>TC227055 similar to GB|AAP21363.1|30102890|BT006555 At4g38290 {Arabidopsis
thaliana;} , partial (42%)
Length = 613
Score = 225 bits (573), Expect = 2e-59
Identities = 117/139 (84%), Positives = 126/139 (90%), Gaps = 1/139 (0%)
Frame = +1
Query: 144 SSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPT-IGNPKGLIDMQWEERYQQLQML 202
SS++SSG+PSVPIS GKS MKLAPVESN VT SSGPT IGNPKGL ++ WEERYQQLQM
Sbjct: 1 SSMNSSGKPSVPISLGKSAMKLAPVESNYVTASSGPTTIGNPKGLKNLHWEERYQQLQMF 180
Query: 203 LRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAK 262
LRKLDQSDQ EY+QML+SLSSVELS+HAVELEKR+IQLSLEEAKELQRVAVLNVLGKS K
Sbjct: 181 LRKLDQSDQEEYIQMLRSLSSVELSKHAVELEKRSIQLSLEEAKELQRVAVLNVLGKSVK 360
Query: 263 KFKAPADHDECSDKLKTSS 281
FKAPADHDECSDKLK S
Sbjct: 361 NFKAPADHDECSDKLKALS 417
>TC227054
Length = 526
Score = 119 bits (297), Expect = 2e-27
Identities = 61/72 (84%), Positives = 63/72 (86%)
Frame = +3
Query: 1 MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS 60
MDLPNRDKQ TV VK TA RDLQNDNKIM TSVGSSS LKDKDP TDSNR+SGTKRPLS
Sbjct: 312 MDLPNRDKQLTVAVK-TALRDLQNDNKIMVPTSVGSSSFLKDKDPGTDSNRVSGTKRPLS 488
Query: 61 DYPVNHHLQQSP 72
DYP+NH LQQSP
Sbjct: 489 DYPLNHQLQQSP 524
>BG651531
Length = 381
Score = 66.2 bits (160), Expect = 2e-11
Identities = 32/47 (68%), Positives = 36/47 (76%), Gaps = 6/47 (12%)
Frame = +3
Query: 111 RQLCSEEETALPK------PQLKEPNVSCFPAFAPFPLTSSISSSGR 151
RQLC EEETA PK PQLKEP SCFPAFAPFP+ SS++SSG+
Sbjct: 240 RQLCCEEETAQPKSQIKEPPQLKEPKASCFPAFAPFPMASSMNSSGK 380
>TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucosidase,
partial (69%)
Length = 2006
Score = 31.6 bits (70), Expect = 0.42
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 10/64 (15%)
Frame = -2
Query: 141 PLTSSISSSGRPSVPISCGKSTMKLAPVESNC----VTTSSGPTI------GNPKGLIDM 190
P+ S+ P+ C K+ + LA +NC V TS GP G+P + ++
Sbjct: 679 PIVRSLEGDNGREFPLFCSKTILSLAASRANC*WAEVHTSLGPRFPYGWVDGSPSNIPNL 500
Query: 191 QWEE 194
W E
Sbjct: 499 IWTE 488
>BF424213 weakly similar to GP|21322711|em pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (6%)
Length = 424
Score = 30.8 bits (68), Expect = 0.72
Identities = 22/66 (33%), Positives = 28/66 (42%), Gaps = 5/66 (7%)
Frame = +1
Query: 108 QPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSS-----ISSSGRPSVPISCGKST 162
QPP + LP P L P++S P+ PFPL S P P S K+T
Sbjct: 187 QPPPSSLNPPPFLLPSPTLTPPSLSTLPS-PPFPLPKP*P*P*PSPPPPPPPPSSSSKTT 363
Query: 163 MKLAPV 168
L P+
Sbjct: 364 QTLHPL 381
>CA851543
Length = 633
Score = 30.8 bits (68), Expect = 0.72
Identities = 26/79 (32%), Positives = 34/79 (42%)
Frame = +3
Query: 124 PQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPTIGN 183
PQ+K F F L +SS S + G ++L ESN TT+ G IG+
Sbjct: 117 PQMKSLLFISFKERQIFFLAIQMSSLILNSTLMILGNFMVELWQRESNGTTTAEGHKIGS 296
Query: 184 PKGLIDMQWEERYQQLQML 202
I QWE + Q Q L
Sbjct: 297 LAMAIGCQWEWKSQ*FQTL 353
>TC220931 similar to UP|Q71QD5 (Q71QD5) Protein phosphatase 2C, partial (11%)
Length = 453
Score = 30.4 bits (67), Expect = 0.94
Identities = 24/99 (24%), Positives = 41/99 (41%)
Frame = -1
Query: 115 SEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVT 174
+ E+ LP EP P P + +SG + G + +L C++
Sbjct: 405 NSEQPQLPSIWNYEPRYR-----TPLPEKRAYPASGASGCSPAAGLTRQQL------CIS 259
Query: 175 TSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVE 213
+ GP G KG+ ++WE++ ++L L Q Q E
Sbjct: 258 RAKGPNSGVRKGVRWVRWEKKSRRLLQQLHSKSQPFQNE 142
>BI699461
Length = 426
Score = 30.0 bits (66), Expect = 1.2
Identities = 18/60 (30%), Positives = 29/60 (48%)
Frame = +1
Query: 86 RKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSS 145
RK E GKC +NP++++ P + EEE L +P + + + F+P SS
Sbjct: 145 RKDNGEDGKCDTRDNPSSSSTNGKPSAV-EEEEVILFRPLARYNSAPLYALFSPHEQISS 321
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 30.0 bits (66), Expect = 1.2
Identities = 26/85 (30%), Positives = 35/85 (40%), Gaps = 5/85 (5%)
Frame = +2
Query: 100 NPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCF---PAFAPFPLTSSISSSGRPSVPI 156
NPT+ PP L S + LP P P +S P+ A P T S SS+ S P
Sbjct: 596 NPTS-----PPTSLASSTFSRLPNPPTPNPPLSGLRPAPSTASTPKTLSPSSTAPTSPPA 760
Query: 157 SC--GKSTMKLAPVESNCVTTSSGP 179
S + + +P + T S P
Sbjct: 761 STPPPRRPARKSPTPTTTFTASPSP 835
>TC225257 similar to UP|Q7XXS4 (Q7XXS4) Thiamine biosynthetic enzyme, partial
(30%)
Length = 587
Score = 29.6 bits (65), Expect = 1.6
Identities = 17/62 (27%), Positives = 27/62 (43%)
Frame = +1
Query: 97 AFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPI 156
+ + P +++ + P + P P PN + FP P P T+S S+ PS
Sbjct: 190 SLQTPNSHSSTESPSHILPASR---PPPNYPHPNKAQFPCP*PHPPTTSSPSNSSPSKNP 360
Query: 157 SC 158
SC
Sbjct: 361 SC 366
>TC206159 similar to UP|SY71_ARATH (Q9SF29) Syntaxin 71 (AtSYP71), partial
(42%)
Length = 520
Score = 29.6 bits (65), Expect = 1.6
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Frame = +1
Query: 209 SDQVEYVQMLQSLSSVELSRHAVELE---KRAIQLSLEEAKELQRVAVLNVLGKSAKKFK 265
+D V +Q +S S + AV + +R LEE +LQR+AV V G S+++F
Sbjct: 283 ADIVALLQKAESASKEKGKASAVAINAEIRRTKARLLEEVPKLQRLAVKKVKGLSSQEFA 462
Query: 266 APAD 269
A D
Sbjct: 463 ARND 474
>TC214199 weakly similar to UP|Q6NLH4 (Q6NLH4) At5g54470, partial (17%)
Length = 878
Score = 29.3 bits (64), Expect = 2.1
Identities = 12/44 (27%), Positives = 23/44 (52%)
Frame = -1
Query: 86 RKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEP 129
R+ E+G CA + ++ +C PRQ+ + E ++P +P
Sbjct: 665 REDTCEVGMCAQNQLCWHSRYCLQPRQILTRGEASVPTKSQPQP 534
>CA785320
Length = 409
Score = 29.3 bits (64), Expect = 2.1
Identities = 23/84 (27%), Positives = 37/84 (43%)
Frame = +3
Query: 101 PTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGK 160
P+ + PP CS+ T+ P + +S +P T++ SS+ RPS P S
Sbjct: 3 PSATSATPPPANPCSK--TSSPSSPVTASALSAWP-------TTATSSTIRPSEPTSKTT 155
Query: 161 STMKLAPVESNCVTTSSGPTIGNP 184
+ P+ +TSS P +P
Sbjct: 156 APSSTPPLTPRLSSTSSPPPNTDP 227
>TC205112 similar to UP|Q93ZP1 (Q93ZP1) AT5g61020/maf19_20, partial (38%)
Length = 1757
Score = 28.9 bits (63), Expect = 2.7
Identities = 16/45 (35%), Positives = 21/45 (46%)
Frame = -1
Query: 108 QPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRP 152
QP C + T LPK PN+S P P LT + + G+P
Sbjct: 1127 QPVHLSCCQ*STVLPKST--GPNISAKPTNCPLVLTENNKNMGQP 999
>TC233627 weakly similar to UP|Q7PKR5 (Q7PKR5) ENSANGP00000023757 (Fragment),
partial (69%)
Length = 905
Score = 28.5 bits (62), Expect = 3.6
Identities = 19/55 (34%), Positives = 25/55 (44%), Gaps = 2/55 (3%)
Frame = +2
Query: 140 FPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTT--SSGPTIGNPKGLIDMQW 192
+PLT + PS SC ST +APV S ++ SSGP+ P W
Sbjct: 59 WPLTCQERTLNAPS---SCSPSTTPMAPVPSTAASSAVSSGPST*TPPAPPSRSW 214
>TC215140 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(91%)
Length = 1885
Score = 28.5 bits (62), Expect = 3.6
Identities = 24/85 (28%), Positives = 35/85 (40%), Gaps = 5/85 (5%)
Frame = +1
Query: 100 NPTNNAFCQPPRQLCSEEETALPKPQLKEP---NVSCFPAFAPFPLTSSISSSGRPSVPI 156
NPT+ PP L S + LP P + P + P+ A P T S SS+ + P
Sbjct: 640 NPTS-----PPTLLASSTSSRLPNPPIPNPP*SGLHPAPSTASTPKTLSPSSTAPTNPPA 804
Query: 157 SC--GKSTMKLAPVESNCVTTSSGP 179
S + + +P + T S P
Sbjct: 805 STPPPRKPARRSPTPTTTSTASPSP 879
>TC217294 similar to UP|Q8SKU2 (Q8SKU2) Tic62 protein precursor, partial
(54%)
Length = 1301
Score = 28.5 bits (62), Expect = 3.6
Identities = 27/103 (26%), Positives = 46/103 (44%), Gaps = 4/103 (3%)
Frame = +2
Query: 83 YVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPL 142
Y+ + ++ + + P N +P ++ +++ETA PKP K+P +P+ +
Sbjct: 452 YISSPKKPDIAAVSVPDPPANVVTVEP--KVATQQETAQPKPVAKQP-------LSPYIV 604
Query: 143 TSSIS--SSGRPSVPISCGKST--MKLAPVESNCVTTSSGPTI 181
+ SS PS P GK T + P S T SS P +
Sbjct: 605 YDDLKPPSSPSPSQP-GGGKPTKISETVPKPSASDTPSSVPGV 730
>TC222248 weakly similar to GB|AAC51925.1|2708305|AF016369 U4/U6 small
nuclear ribonucleoprotein hPrp4 {Homo sapiens;} ,
partial (7%)
Length = 747
Score = 28.1 bits (61), Expect = 4.6
Identities = 20/67 (29%), Positives = 32/67 (46%)
Frame = +3
Query: 181 IGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQL 240
+G P L + ER +L+M++ KLD Q+E + A E E+ A
Sbjct: 579 LGEPITLFGEREMERRDRLRMIMAKLDAEGQLEKLM------------KAHEEEEAAASA 722
Query: 241 SLEEAKE 247
S++EA+E
Sbjct: 723 SIDEAEE 743
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.127 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,100,769
Number of Sequences: 63676
Number of extensions: 186420
Number of successful extensions: 938
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 933
length of query: 281
length of database: 12,639,632
effective HSP length: 96
effective length of query: 185
effective length of database: 6,526,736
effective search space: 1207446160
effective search space used: 1207446160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0117b.7


