

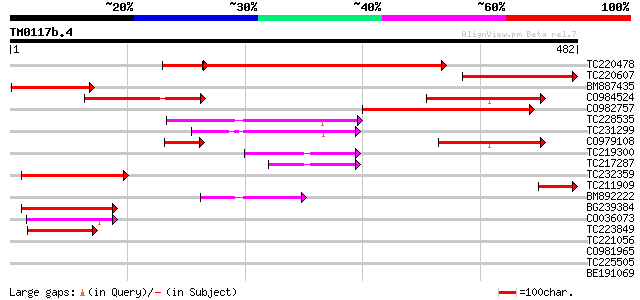
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.4
(482 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220478 383 e-119
TC220607 similar to UP|GLSA_PSESM (Q882Y4) Probable glutaminase ... 172 2e-43
BM887435 123 2e-28
CO984524 120 2e-27
CO982757 115 4e-26
TC228535 88 1e-17
TC231299 84 2e-16
CO979108 77 1e-14
TC219300 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial... 77 2e-14
TC217287 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial... 67 2e-11
TC232359 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial... 64 1e-10
TC211909 UP|Q76PT8 (Q76PT8) D5L protein, partial (16%) 60 2e-09
BM892222 57 1e-08
BG239384 56 3e-08
CO036073 49 7e-06
TC223849 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial... 48 1e-05
TC221056 weakly similar to UP|Q93ZL1 (Q93ZL1) AT5g60720/mup24_13... 39 0.007
CO981965 34 0.17
TC225505 similar to UP|PSA2_ARATH (O23708) Proteasome subunit al... 31 1.4
BE191069 30 1.8
>TC220478
Length = 801
Score = 383 bits (983), Expect(2) = e-119
Identities = 188/207 (90%), Positives = 195/207 (93%)
Frame = +1
Query: 165 MIIKSFTSIMAVILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDEL 224
M+ K+FT+I+AVILEAFGVYCEGEFK+GCGYPY+AVVLNFSQSWALYCLVQFYT TKDEL
Sbjct: 181 MLTKAFTAILAVILEAFGVYCEGEFKVGCGYPYMAVVLNFSQSWALYCLVQFYTDTKDEL 360
Query: 225 AHIKPLAKFLTFKSIVFLTWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGI 284
AHIKPLAKFLTFKSIVFLTWWQGVAIALL TFGL KSPIAQGLQ KSSVQDFIICIEMGI
Sbjct: 361 AHIKPLAKFLTFKSIVFLTWWQGVAIALLSTFGLIKSPIAQGLQFKSSVQDFIICIEMGI 540
Query: 285 ASIVHLYVFPAKPYELMGDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDA 344
ASIVHLYVFPAKPYE MGDR GS+SVLGDYSADCPLDPDEIRDSERPTKLRLPT DVD
Sbjct: 541 ASIVHLYVFPAKPYERMGDRFSGSVSVLGDYSADCPLDPDEIRDSERPTKLRLPTPDVDT 720
Query: 345 KSGMTIRESVRDVVIGGSGYIVKDVKF 371
KSGMTIRESVRDVVIGG GYI KDVKF
Sbjct: 721 KSGMTIRESVRDVVIGGGGYIEKDVKF 801
Score = 65.9 bits (159), Expect(2) = e-119
Identities = 29/39 (74%), Positives = 31/39 (79%)
Frame = +3
Query: 131 KGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKS 169
K TVNHPFPL YFLKPW L R FYQI+KFGIVQY+ S
Sbjct: 6 KATVNHPFPLNYFLKPWKLGRAFYQIIKFGIVQYVCYAS 122
>TC220607 similar to UP|GLSA_PSESM (Q882Y4) Probable glutaminase , partial
(6%)
Length = 805
Score = 172 bits (437), Expect = 2e-43
Identities = 82/97 (84%), Positives = 89/97 (91%)
Frame = +2
Query: 386 RFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGIDDPLLNGSISDSGISRAK 445
R NEKLHRIS+N+KKHDK+ RR KDDSCIASSSPARRVIRGIDDPLLNGS+SDSG+ R K
Sbjct: 2 RVNEKLHRISENMKKHDKNGRRTKDDSCIASSSPARRVIRGIDDPLLNGSVSDSGMLRVK 181
Query: 446 KHRRKSGYTSAESGGESSSDQSYGAYQVRGHRWVTKE 482
KHRRKSGYTSAESGGESSSDQ +G YQ+RG RWVTKE
Sbjct: 182 KHRRKSGYTSAESGGESSSDQGFGGYQIRGRRWVTKE 292
>BM887435
Length = 428
Score = 123 bits (308), Expect = 2e-28
Identities = 59/71 (83%), Positives = 65/71 (91%)
Frame = +2
Query: 2 IIIQLLHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSI 61
I+IQL +PP WAT++A+ FLL+TLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYS
Sbjct: 215 ILIQLFSSPPAWATLIAAAFLLLTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSF 394
Query: 62 ESFVSLVNPSI 72
ES VSLVNPSI
Sbjct: 395 ESCVSLVNPSI 427
>CO984524
Length = 685
Score = 120 bits (300), Expect = 2e-27
Identities = 54/103 (52%), Positives = 75/103 (72%)
Frame = -2
Query: 64 FVSLVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLL 123
F+SL++ S + +C ++RDCYE+FA+YCF RYL+ACLGGE++TI+FME + S TPLL
Sbjct: 654 FLSLLDSSAAFNCEVIRDCYEAFALYCFERYLIACLGGEDKTIQFMESMSLTESSTPLLK 475
Query: 124 HHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMI 166
+++ G V HPFP+ FL+ W L FYQ VK GIVQY++
Sbjct: 474 EAYAY---GVVEHPFPVNCFLRDWYLGPDFYQSVKIGIVQYIV 355
Score = 82.0 bits (201), Expect = 5e-16
Identities = 44/105 (41%), Positives = 65/105 (61%), Gaps = 4/105 (3%)
Frame = -2
Query: 355 RDVVIGGSGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRR---RIKDD 411
+ V IG YIV D+KFTV VEPVE+GI + N+ H+IS+N+K+H+ +RR ++KDD
Sbjct: 390 QSVKIGIVQYIVDDMKFTVSHVVEPVERGIAKINKTFHQISENVKRHEDERRKSTKVKDD 211
Query: 412 SCIASSSPARRVIRGIDDPLLNGSISDSGI-SRAKKHRRKSGYTS 455
S + + R + D L GS+SDSG+ SR ++H + S
Sbjct: 210 SYLVALRSWRSEFSDVHDRLGEGSVSDSGLPSRKRQHLQSKASAS 76
>CO982757
Length = 659
Score = 115 bits (288), Expect = 4e-26
Identities = 61/148 (41%), Positives = 91/148 (61%), Gaps = 2/148 (1%)
Frame = -1
Query: 301 MGDRHPGSISVLGDY-SADCPLDPDEIRDSERPTKLRLPT-TDVDAKSGMTIRESVRDVV 358
MG+R +++V+ DY S PLDP+E++DS+R T+ L + K+ M +SVRDVV
Sbjct: 656 MGERCIRNVAVMDDYASLGSPLDPEEVQDSQRSTRTWLGAHNNQREKNPMKFTQSVRDVV 477
Query: 359 IGGSGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSS 418
+G IV D+KFTV VEPVE+GI + N+ H+IS+N+K+H++ R KDD +
Sbjct: 476 VGSGEIIVDDMKFTVSHVVEPVERGIAKINKTFHQISENVKRHEQRTRNTKDDCYLVPLR 297
Query: 419 PARRVIRGIDDPLLNGSISDSGISRAKK 446
+ D + GS+SDSG+SR K+
Sbjct: 296 TQMSEFSDVHDTMGEGSVSDSGMSRVKR 213
>TC228535
Length = 980
Score = 87.8 bits (216), Expect = 1e-17
Identities = 56/170 (32%), Positives = 87/170 (50%), Gaps = 3/170 (1%)
Frame = +1
Query: 134 VNHPFPLKYFLKPWI-LTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLG 192
++H FP+ F L + +++K Q+++I+ SI+ + L+ VY +
Sbjct: 181 IHHSFPMTLFQPHTTRLDHKTLKLLKNWTWQFVVIRPVCSILMITLQYLDVYPTWVSWIN 360
Query: 193 CGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIAL 252
V+LN S S ALY LV FY V EL KPLAKFL K IVF +WQG+ + L
Sbjct: 361 ------TVILNISVSLALYSLVVFYHVFSKELEPHKPLAKFLCIKGIVFFCFWQGIVLDL 522
Query: 253 LYTFGLFKSPIA--QGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYEL 300
L G+ +S + + + Q+ ++C+EM SI Y + A PY++
Sbjct: 523 LAALGIIRSRYSWLAVERIEEGYQNLLVCVEMVFFSIYQQYAYSAAPYKV 672
>TC231299
Length = 517
Score = 83.6 bits (205), Expect = 2e-16
Identities = 50/146 (34%), Positives = 82/146 (55%), Gaps = 2/146 (1%)
Frame = +1
Query: 155 QIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLV 214
+++K+ Q+++++ S++ + L+ G+Y L + +VLN S S ALY LV
Sbjct: 49 KLLKYWTWQFVVVRPVCSVLMIALQLVGLYPTW---LSWAF---TIVLNISVSLALYSLV 210
Query: 215 QFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTFGLFKSPIAQ--GLQSKSS 272
FY V ELA KPLAKFL K IVF +WQG+ + LL G+ +S + + +
Sbjct: 211 VFYHVFAKELAPHKPLAKFLCIKGIVFFCFWQGMLLELLAATGVIQSRHLRLDVEHIEEA 390
Query: 273 VQDFIICIEMGIASIVHLYVFPAKPY 298
+Q+ ++C+EM I S++ Y + PY
Sbjct: 391 MQNILVCLEMVIFSVLQQYAYHPAPY 468
>CO979108
Length = 747
Score = 77.4 bits (189), Expect = 1e-14
Identities = 40/95 (42%), Positives = 60/95 (63%), Gaps = 4/95 (4%)
Frame = -1
Query: 365 IVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRR---RIKDDSCIASSSPAR 421
IV D+KFTV VEPVE+GI + N+ H+IS+N+K+H+ +RR ++KDDS + + R
Sbjct: 360 IVDDMKFTVSHVVEPVERGIAKINKTFHQISENVKRHEDERRKSTKVKDDSYLVALRSWR 181
Query: 422 RVIRGIDDPLLNGSISDSGI-SRAKKHRRKSGYTS 455
+ D L GS+SDSG+ SR ++H + S
Sbjct: 180 SEFSDVHDRLGEGSVSDSGLPSRKRQHLQSKASAS 76
Score = 46.2 bits (108), Expect = 3e-05
Identities = 20/34 (58%), Positives = 23/34 (66%)
Frame = -3
Query: 132 GTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYM 165
G V HPFP+ FL+ W L FYQ VK GIVQY+
Sbjct: 730 GVVEHPFPVNCFLRDWYLGPDFYQSVKIGIVQYV 629
>TC219300 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial (30%)
Length = 511
Score = 76.6 bits (187), Expect = 2e-14
Identities = 40/99 (40%), Positives = 54/99 (54%)
Frame = +2
Query: 200 VVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTFGLF 259
++ S + ALY L FY KD L P+ KF+ KS+VFLT+WQGV F
Sbjct: 2 IIYMISYTMALYVLALFYVACKDLLQPFNPVPKFIIIKSVVFLTYWQGVLF-----FLAA 166
Query: 260 KSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPY 298
KS + + +Q+FIIC+EM +A+ H Y FP K Y
Sbjct: 167 KSGFIEDADEAALLQNFIICVEMLVAAAGHFYAFPYKEY 283
>TC217287 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial (49%)
Length = 1093
Score = 67.0 bits (162), Expect = 2e-11
Identities = 33/78 (42%), Positives = 45/78 (57%)
Frame = +3
Query: 221 KDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICI 280
KD L P+ KF+ KS+VFLT+WQGV + F KS + + +QDF IC+
Sbjct: 9 KDLLQPFNPVPKFIIIKSVVFLTYWQGVLV-----FLAAKSEFVKDADEAALLQDFFICV 173
Query: 281 EMGIASIVHLYVFPAKPY 298
EM +A++ H Y FP K Y
Sbjct: 174 EMLVAAVGHFYAFPYKEY 227
>TC232359 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial (37%)
Length = 719
Score = 64.3 bits (155), Expect = 1e-10
Identities = 31/91 (34%), Positives = 55/91 (60%)
Frame = +3
Query: 11 PVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNP 70
P++ IVA + +ALS+ +++HL Y P Q+F++ ++ MVP Y++ SF+SLV P
Sbjct: 246 PIFLYIVAFICTCGAIALSLLHIYKHLLNYTEPTYQRFIVRIVFMVPVYALMSFLSLVLP 425
Query: 71 SISVDCGILRDCYESFAMYCFGRYLVACLGG 101
S+ +R+ YE++ +Y F + +GG
Sbjct: 426 QGSIYFNSIREIYEAWVIYNFLSLCLEWVGG 518
>TC211909 UP|Q76PT8 (Q76PT8) D5L protein, partial (16%)
Length = 562
Score = 60.5 bits (145), Expect = 2e-09
Identities = 26/33 (78%), Positives = 30/33 (90%)
Frame = +3
Query: 450 KSGYTSAESGGESSSDQSYGAYQVRGHRWVTKE 482
KSGYTSAE GGES+S+QS+G YQ+RG RWVTKE
Sbjct: 3 KSGYTSAEVGGESTSEQSFGGYQIRGRRWVTKE 101
>BM892222
Length = 422
Score = 57.4 bits (137), Expect = 1e-08
Identities = 37/90 (41%), Positives = 47/90 (52%)
Frame = +1
Query: 163 QYMIIKSFTSIMAVILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKD 222
Q+++I+ SI+ + L+ VY V+LN S S ALY LV FY V
Sbjct: 34 QFVVIRPVCSILMITLQYLEVYPTWVSWTN------TVILNISVSLALYSLVVFYHVFSK 195
Query: 223 ELAHIKPLAKFLTFKSIVFLTWWQGVAIAL 252
EL KPLAKFL K IVF +WQ + AL
Sbjct: 196 ELEPHKPLAKFLCIKGIVFFCFWQVFSYAL 285
>BG239384
Length = 430
Score = 56.2 bits (134), Expect = 3e-08
Identities = 24/81 (29%), Positives = 50/81 (61%)
Frame = +2
Query: 11 PVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNP 70
P++ +VA + + +AL++ ++ HL +Y P Q++++ +I MVP Y+++SF+ L P
Sbjct: 167 PLFFHVVAFICTVAAIALAVLHIYRHLLSYTEPTYQRYIVRIIFMVPGYALKSFLVLCIP 346
Query: 71 SISVDCGILRDCYESFAMYCF 91
+ +R+ YE++ +Y F
Sbjct: 347 GSPIYFNSIREVYEAWVIYNF 409
>CO036073
Length = 385
Score = 48.5 bits (114), Expect = 7e-06
Identities = 29/82 (35%), Positives = 48/82 (58%), Gaps = 5/82 (6%)
Frame = -3
Query: 15 TIVASVFLLM-TLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSIS 73
T++ S F M ++ + LL +HL +KNP+EQK +I +ILM P Y+ SFV L++ S
Sbjct: 362 TVLGSAFCAMLSMHFTSQLLSQHLFYWKNPKEQKAIIIIILMAPIYAAVSFVGLLDIRGS 183
Query: 74 VD----CGILRDCYESFAMYCF 91
+ +++CYE+ + F
Sbjct: 182 KEFFTFLESVKECYEALVIAKF 117
>TC223849 similar to UP|Q9SFF8 (Q9SFF8) F2O10.10 protein, partial (15%)
Length = 440
Score = 47.8 bits (112), Expect = 1e-05
Identities = 22/59 (37%), Positives = 37/59 (62%)
Frame = +2
Query: 16 IVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSISV 74
IVA + +ALS+ +++HL Y P Q+F++ ++ MVP Y++ SF+SL P S+
Sbjct: 260 IVAFICTCGAIALSLLHIYKHLLNYTEPTYQRFIVRIVFMVPVYALMSFLSLFLPQGSI 436
>TC221056 weakly similar to UP|Q93ZL1 (Q93ZL1) AT5g60720/mup24_130, partial
(15%)
Length = 1022
Score = 38.5 bits (88), Expect = 0.007
Identities = 19/31 (61%), Positives = 20/31 (64%)
Frame = +2
Query: 216 FYTVTKDELAHIKPLAKFLTFKSIVFLTWWQ 246
FY V ELA KPLAKFL K IVF +WQ
Sbjct: 5 FYHVFAKELAPHKPLAKFLCIKGIVFFCFWQ 97
>CO981965
Length = 843
Score = 33.9 bits (76), Expect = 0.17
Identities = 21/50 (42%), Positives = 33/50 (66%), Gaps = 5/50 (10%)
Frame = -2
Query: 252 LLYTFGLFKSPIA--QGLQSKS-SVQDF--IICIEMGIASIVHLYVFPAK 296
+ + F LF+S +A QG S S S +DF + CI+ GI+SI+H Y+F ++
Sbjct: 203 ITFRFKLFRSSVALPQGSFSYSRSSKDF*SLPCIDKGISSILHFYIFSSE 54
>TC225505 similar to UP|PSA2_ARATH (O23708) Proteasome subunit alpha type 2
(20S proteasome alpha subunit B) , partial (46%)
Length = 632
Score = 30.8 bits (68), Expect = 1.4
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 4/53 (7%)
Frame = -3
Query: 230 LAKFLTFKSIVFLTWWQGVAIA----LLYTFGLFKSPIAQGLQSKSSVQDFII 278
L F+T + + W +A + T LFK PI ++ S QDF I
Sbjct: 495 LKMFMTLEKVYLKACWSSQRVARKFTIAITMSLFKPPIKPNCETTSGTQDFFI 337
>BE191069
Length = 577
Score = 30.4 bits (67), Expect = 1.8
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = +2
Query: 260 KSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPY 298
KS + + +Q+FII +EM +A++ H Y F K Y
Sbjct: 11 KSGFIEDADEAALLQNFII*VEMLVAAVGHFYAFTYKEY 127
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,612,428
Number of Sequences: 63676
Number of extensions: 325607
Number of successful extensions: 2015
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1982
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2002
length of query: 482
length of database: 12,639,632
effective HSP length: 101
effective length of query: 381
effective length of database: 6,208,356
effective search space: 2365383636
effective search space used: 2365383636
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0117b.4


