

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.11
(669 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
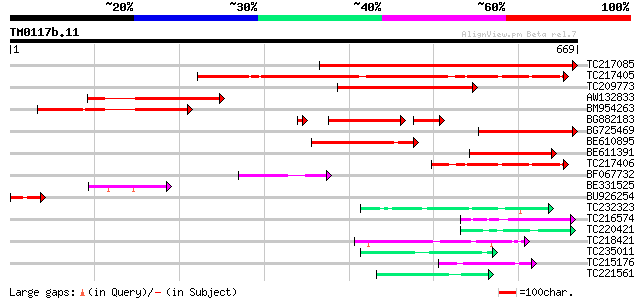
Score E
Sequences producing significant alignments: (bits) Value
TC217085 502 e-142
TC217405 similar to UP|Q9FLL9 (Q9FLL9) Gb|AAB80666.1, partial (20%) 361 e-100
TC209773 similar to UP|Q9LW19 (Q9LW19) Gb|AAB80666.1, partial (3%) 241 8e-64
AW132833 211 6e-55
BM954263 192 3e-49
BG882183 136 6e-46
BG725469 145 4e-35
BE610895 138 9e-33
BE611391 137 2e-32
TC217406 similar to UP|Q9FLL9 (Q9FLL9) Gb|AAB80666.1, partial (9%) 137 2e-32
BF067732 similar to GP|2707600|gb|A triple LIM domain protein {H... 73 4e-13
BE331525 69 9e-12
BU926254 53 5e-07
TC232323 UP|PUR1_SOYBN (P52418) Amidophosphoribosyltransferase, ... 45 1e-04
TC216574 similar to UP|RK4_SPIOL (O49937) 50S ribosomal protein ... 45 1e-04
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 45 1e-04
TC218421 similar to GB|AAQ65192.1|34365761|BT010569 At3g08670 {A... 44 3e-04
TC235011 weakly similar to UP|Q8V7F2 (Q8V7F2) ORF1 (Fragment), p... 43 4e-04
TC215176 similar to GB|BAC42264.1|26450296|AK117608 GPI-anchored... 43 4e-04
TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate d... 42 7e-04
>TC217085
Length = 1262
Score = 502 bits (1292), Expect = e-142
Identities = 257/305 (84%), Positives = 276/305 (90%), Gaps = 1/305 (0%)
Frame = +3
Query: 366 GSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTN 425
GSQSAPLFADNKPD+SEKLRQMRPSLSRKFSSYVLPTPVDAKSS SSGSNNPKPSKMQ N
Sbjct: 6 GSQSAPLFADNKPDASEKLRQMRPSLSRKFSSYVLPTPVDAKSSTSSGSNNPKPSKMQAN 185
Query: 426 SNEATTNLWHSSPLEQKKHET-IRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLL 484
NE T NLWHSSPLEQKK E I DEFS VR+AQSVLKESNSNTA+ RLPPPL+D LL
Sbjct: 186 LNEPTKNLWHSSPLEQKKQEKDIGDEFSGSIVRSAQSVLKESNSNTASMRLPPPLIDSLL 365
Query: 485 SSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKV 544
SSN DY+SAYSKKIKRHAFSGPLTSNP PT+PVS++SVQ+FSGPLLP PIPQPPSSSPK
Sbjct: 366 SSNRDYISAYSKKIKRHAFSGPLTSNPGPTKPVSIESVQLFSGPLLPAPIPQPPSSSPKA 545
Query: 545 SPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISS 604
SPSASPT+++SPKISELHELPRPPT+FPSNSRLLGLVG+SGPLV RGQKV+A NNLV SS
Sbjct: 546 SPSASPTLMTSPKISELHELPRPPTSFPSNSRLLGLVGHSGPLVSRGQKVTA-NNLVASS 722
Query: 605 AASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSR 664
AASPLP+PPQAMARSFSIPSSGARV ALH LESSH SSISE+IASPPL PI+LS+SR
Sbjct: 723 AASPLPMPPQAMARSFSIPSSGARVAALHMPRTLESSHRSSISENIASPPLMPISLSSSR 902
Query: 665 PSSDG 669
PS DG
Sbjct: 903 PSLDG 917
>TC217405 similar to UP|Q9FLL9 (Q9FLL9) Gb|AAB80666.1, partial (20%)
Length = 1652
Score = 361 bits (926), Expect = e-100
Identities = 222/445 (49%), Positives = 291/445 (64%), Gaps = 7/445 (1%)
Frame = +1
Query: 222 LFVFRLKSLKQGQSRSLLTQAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHIDYHFN 281
L FRLKSLKQGQSRSLLTQAARHHAAQL FF+K +KSLE V+PHV+ + E+QHIDY F+
Sbjct: 10 LCAFRLKSLKQGQSRSLLTQAARHHAAQLNFFRKGLKSLEAVDPHVRMIAERQHIDYQFS 189
Query: 282 GLEEEDGDEGDEGYEDDDGYDENDDGELSFDYGQIEQ-EQDVSTSRNSMELDQVELTLPR 340
GL E+DG E D +D + +D + GELSFDY +Q VSTS NS E+++ + R
Sbjct: 190 GL-EDDGGENDNN-DDGNDFDVIEGGELSFDYRANKQGPYIVSTSPNSAEVEESGRSYIR 363
Query: 341 GSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVL 400
ST E A+ +L+K Q + GS SAP+ A+ K D +EK+RQ+ + K ++YVL
Sbjct: 364 ASTPETAETSLEKNQGDFKVSNRVGGSYSAPILAEKKFDPAEKVRQLLAFSAAKSNAYVL 543
Query: 401 PTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHE-TIRDEFSSPTVRNA 459
PTPV+ K + +S S T+++ + NLWHSSPL++KK+E + + S PT+ A
Sbjct: 544 PTPVNIKETKTS-------SAPHTSASGSLHNLWHSSPLDEKKNEKDVDSKLSEPTIPRA 702
Query: 460 QSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAY-SKKIKRHAFSGPLTSNPMPTRPVS 518
SVLKESNS+T T+LP P DGL D SA +KKIKR AFSGPLT+ P+ +PV
Sbjct: 703 HSVLKESNSDTTYTQLPRPSADGLSPPQVDLFSATDTKKIKRQAFSGPLTNKPLSVKPV- 879
Query: 519 VDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLL 578
SG P+PQP SSPK SP+ASP +VSSP+ISELHELPRPP N S R
Sbjct: 880 -------SGGFPRLPVPQP--SSPKASPNASPPLVSSPRISELHELPRPPGNQSSKPR-- 1026
Query: 579 GLVGYSGPLVPRGQKVSAPNNL--VISSAASPLPIPPQAMARSFSIPSSGARVTALHSSS 636
VG+S PLV + + + N V+ SAASPLP PP ++RSFSIPSSG RV AL+ S+
Sbjct: 1027--VGHSAPLVLKNPEAAMTNKFPSVVFSAASPLPTPP--VSRSFSIPSSGQRVVALNVSN 1194
Query: 637 A--LESSHISSISEDIASPPLTPIA 659
L++ +S + + + SPPLTP++
Sbjct: 1195TKYLDTPQVSKVDKAV-SPPLTPMS 1266
>TC209773 similar to UP|Q9LW19 (Q9LW19) Gb|AAB80666.1, partial (3%)
Length = 506
Score = 241 bits (615), Expect = 8e-64
Identities = 125/168 (74%), Positives = 140/168 (82%), Gaps = 3/168 (1%)
Frame = +1
Query: 388 RPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETI 447
R +LSRKF+SYVLPTPVD KSSIS S+N PSK++TN NE NLWHSSPLE+KK+E I
Sbjct: 1 RQTLSRKFNSYVLPTPVDGKSSISLRSSNQVPSKIKTNLNEPVKNLWHSSPLEKKKYENI 180
Query: 448 RDE--FSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSG 505
+ FS P VR AQSVLKESNSNTA +RLPPPL+DG LSSNHDY++AYSKKIKRHAFSG
Sbjct: 181 FGDGGFSGPDVRTAQSVLKESNSNTAYSRLPPPLIDGNLSSNHDYITAYSKKIKRHAFSG 360
Query: 506 PLTSNPMPTR-PVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTI 552
PL SN PT+ PVSV SVQ+FSGPLL T IPQPPSSSPKVSPSASP +
Sbjct: 361 PLVSNAWPTKPPVSVKSVQLFSGPLLRTSIPQPPSSSPKVSPSASPPL 504
>AW132833
Length = 385
Score = 211 bits (538), Expect = 6e-55
Identities = 115/161 (71%), Positives = 119/161 (73%)
Frame = +2
Query: 93 GKVLLMLGKIQFKLQKLIDKYVCISLLIPSLYFIFCFCFNFITNFSVFLFFQFAQRSHII 152
GK+LLMLGKIQFKL KLID Y RSHI
Sbjct: 5 GKLLLMLGKIQFKLHKLIDHY----------------------------------RSHIF 82
Query: 153 QTITIPSESLLNELRIVEEMKRQCDEKRDVYEYMATRFRERGRSKGGKTETFSLQQLQTA 212
QTITIPSESLLNELRIVEEMKRQCDEKRD+Y+YM TR+RE G SKGGK ETFSLQQLQTA
Sbjct: 83 QTITIPSESLLNELRIVEEMKRQCDEKRDLYDYMVTRYREGGXSKGGKGETFSLQQLQTA 262
Query: 213 RDEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQLCFF 253
DEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQLCFF
Sbjct: 263 HDEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQLCFF 385
>BM954263
Length = 454
Score = 192 bits (489), Expect = 3e-49
Identities = 105/183 (57%), Positives = 128/183 (69%)
Frame = +3
Query: 33 QLEELAQATRDMQDMRDCYDTLLSAAAATASSAYEFAESLRDMGSCLLEKTALNDHEEET 92
+ +ELA A +DMQDMRDCYD+LL+AAAAT +SA+EFAESL+DMG+CLLEKTALND +EE+
Sbjct: 9 KFDELALAAKDMQDMRDCYDSLLAAAAATQNSAHEFAESLQDMGTCLLEKTALND-DEES 185
Query: 93 GKVLLMLGKIQFKLQKLIDKYVCISLLIPSLYFIFCFCFNFITNFSVFLFFQFAQRSHII 152
GK L MLG +Q +LQKL+D Y RSHI+
Sbjct: 186 GKFLGMLGSVQLELQKLVDSY----------------------------------RSHIV 263
Query: 153 QTITIPSESLLNELRIVEEMKRQCDEKRDVYEYMATRFRERGRSKGGKTETFSLQQLQTA 212
TIT PSESLLNELR VE+MKRQCDEKR+VYEYM + +E+G+SK GK E+F+LQQLQ A
Sbjct: 264 LTITNPSESLLNELRTVEDMKRQCDEKRNVYEYMIAQQKEKGKSKSGKGESFTLQQLQAA 443
Query: 213 RDE 215
E
Sbjct: 444 HAE 452
>BG882183
Length = 420
Score = 136 bits (342), Expect(3) = 6e-46
Identities = 70/92 (76%), Positives = 77/92 (83%), Gaps = 1/92 (1%)
Frame = +2
Query: 377 KPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHS 436
K ++SEKLRQMRPSLSRKFSSYVL TPVDAKSS SSGSNNPKPS MQ N E T NLWHS
Sbjct: 32 KLNASEKLRQMRPSLSRKFSSYVLATPVDAKSSTSSGSNNPKPSNMQANLREPTKNLWHS 211
Query: 437 SPLEQKKHE-TIRDEFSSPTVRNAQSVLKESN 467
SPLEQKKH+ + DEFS +R+AQS+LKESN
Sbjct: 212 SPLEQKKHDKDMGDEFSGSIIRSAQSLLKESN 307
Score = 65.5 bits (158), Expect(3) = 6e-46
Identities = 31/37 (83%), Positives = 32/37 (85%)
Frame = +1
Query: 477 PPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMP 513
PP VD LLSSN DY+SAYSKKIKRHAFSGPLTSN P
Sbjct: 310 PPSVDSLLSSNRDYISAYSKKIKRHAFSGPLTSNLGP 420
Score = 22.3 bits (46), Expect(3) = 6e-46
Identities = 10/12 (83%), Positives = 10/12 (83%)
Frame = +3
Query: 340 RGSTAEAAKENL 351
RGS AEA KENL
Sbjct: 3 RGSIAEAIKENL 38
>BG725469
Length = 362
Score = 145 bits (367), Expect = 4e-35
Identities = 80/116 (68%), Positives = 91/116 (77%)
Frame = +2
Query: 554 SSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPP 613
+SPKISELHELP PT+FPSNSRLLGLVG+SGPLV RG+K NLV SSAA L +PP
Sbjct: 2 ASPKISELHELPTAPTSFPSNSRLLGLVGHSGPLVFRGEK*LL-FNLVASSAAYTLTMPP 178
Query: 614 QAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSSDG 669
QAMAR+FSIPSSGARV LH LES SSIS+++ASPPL PI+L +SRP DG
Sbjct: 179 QAMARTFSIPSSGARVAXLHMPRTLESCXRSSISDNMASPPLMPISLXSSRPLLDG 346
>BE610895
Length = 376
Score = 138 bits (347), Expect = 9e-33
Identities = 75/128 (58%), Positives = 91/128 (70%), Gaps = 2/128 (1%)
Frame = +2
Query: 357 NLFSFRVRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNN 416
N FSF+V++ SQSAPLF DNK DSSEKLRQMR + K + YV PTP+ A SSIS S+N
Sbjct: 2 NSFSFKVKSASQSAPLFVDNKRDSSEKLRQMRTTGCWKCNEYVQPTPLKATSSISMRSSN 181
Query: 417 PKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDE--FSSPTVRNAQSVLKESNSNTATTR 474
P +++TN N+ NLWHSSPLEQKK+E I + S P NAQ+ L ES SN A +R
Sbjct: 182 QVPLRIKTNLNDPMKNLWHSSPLEQKKYENIFGDGGLSGP---NAQTALNESYSNAAYSR 352
Query: 475 LPPPLVDG 482
LPPPL+DG
Sbjct: 353 LPPPLIDG 376
>BE611391
Length = 313
Score = 137 bits (345), Expect = 2e-32
Identities = 69/103 (66%), Positives = 83/103 (79%)
Frame = +3
Query: 543 KVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVI 602
KVSPSASP + SSPKI+ELHELPRPPT PSNS+ LGLVG+SGPLV RGQ++SAPN +
Sbjct: 3 KVSPSASPPLSSSPKINELHELPRPPTISPSNSKFLGLVGHSGPLVSRGQQLSAPNYVAT 182
Query: 603 SSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISS 645
S+A SPLP+PP AMARSFSIP+ G +VT LH S E+ + S+
Sbjct: 183 SNAPSPLPMPPSAMARSFSIPNIGDKVTELHESRPEEAPNKST 311
>TC217406 similar to UP|Q9FLL9 (Q9FLL9) Gb|AAB80666.1, partial (9%)
Length = 840
Score = 137 bits (344), Expect = 2e-32
Identities = 83/166 (50%), Positives = 108/166 (65%), Gaps = 4/166 (2%)
Frame = +1
Query: 498 IKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPK 557
+KR AFSGPLT+ P+ +PVS G P+PQP SSPK SP+ASP +VSSP+
Sbjct: 1 VKRQAFSGPLTNKPLSVKPVS--------GGFSRLPVPQP--SSPKASPNASPPLVSSPR 150
Query: 558 ISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNL--VISSAASPLPIPPQA 615
ISELHELPRPP N S R VG+S PLV + + + N V+ AASPLP PP
Sbjct: 151 ISELHELPRPPGNQSSKPR----VGHSAPLVLKNPEAAMTNKFPSVVLRAASPLPTPP-- 312
Query: 616 MARSFSIPSSGARVTALHSSSA--LESSHISSISEDIASPPLTPIA 659
++RSFSIPSSG RV AL+ S+ L++ +S + + + SPPLTP++
Sbjct: 313 VSRSFSIPSSGQRVVALNVSNTKYLDTPQVSKVDKAV-SPPLTPMS 447
>BF067732 similar to GP|2707600|gb|A triple LIM domain protein {Homo
sapiens}, partial (6%)
Length = 284
Score = 73.2 bits (178), Expect = 4e-13
Identities = 47/111 (42%), Positives = 60/111 (53%), Gaps = 2/111 (1%)
Frame = +3
Query: 271 TEQQHIDYHFNGLEEEDGDEGDEG--YEDDDGYDENDDGELSFDYGQIEQEQDVSTSRNS 328
T+Q HIDYHF G+E ED DE ++G DD+ D++ DG L+F Y + E DV N
Sbjct: 3 TDQHHIDYHFIGIEGEDEDEDEDGDVDHDDESXDDDYDGVLNFXYTKNEC*HDVPIFWN* 182
Query: 329 MELDQVELTLPRGSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPD 379
M KE LD+L+RN FSF+V + SQSA LF D D
Sbjct: 183 M------------------KE*LDRLRRNSFSFKVXSXSQSAXLFVDYXRD 281
>BE331525
Length = 397
Score = 68.6 bits (166), Expect = 9e-12
Identities = 47/131 (35%), Positives = 66/131 (49%), Gaps = 33/131 (25%)
Frame = +3
Query: 94 KVLLMLGKIQFKLQKLIDKYVC---------------ISLLIPSLYFIFCFCFNFITNFS 138
K+LLMLGKIQFKL KLID YV +SL + + F +F++ FS
Sbjct: 3 KLLLMLGKIQFKLHKLIDHYVTSYFLLSILSLFLSLSLSLSLSLSLSLSSFHHSFLSLFS 182
Query: 139 VFLFFQ-----------------FAQRSHIIQTITIPSESLLNE-LRIVEEMKRQCDEKR 180
V ++ + F S ++ PS + ++ V+EMKRQCDEKR
Sbjct: 183 VLIYSRQLPFPQSLS*MNFELSRFPSLSITLRLFFYPSTLITSDHFFFVQEMKRQCDEKR 362
Query: 181 DVYEYMATRFR 191
D+Y+YM TR+R
Sbjct: 363 DLYDYMVTRYR 395
>BU926254
Length = 363
Score = 52.8 bits (125), Expect = 5e-07
Identities = 29/42 (69%), Positives = 33/42 (78%)
Frame = +1
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATR 42
MKSSLKKLRGL LHNH K SK+I PLGQL+ELA+AT+
Sbjct: 166 MKSSLKKLRGLALHNH----KLVSSKSIRPLGQLDELARATQ 279
>TC232323 UP|PUR1_SOYBN (P52418) Amidophosphoribosyltransferase, chloroplast
precursor (Glutamine phosphoribosylpyrophosphate
amidotransferase) (ATASE) (GPAT) , partial (72%)
Length = 1231
Score = 45.1 bits (105), Expect = 1e-04
Identities = 59/234 (25%), Positives = 94/234 (39%), Gaps = 6/234 (2%)
Frame = +1
Query: 414 SNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATT 473
+ NP +K + +N L H++ +K +T+ SS T + V + + T
Sbjct: 31 TTNPFQTKTLSLTNSLPLCLSHAT----QKTKTL---VSSSTTKTKSPVKSAAWWASTAT 189
Query: 474 RLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTP 533
R PP + + S +KK+ S TSNP+P S S S P P P
Sbjct: 190 RKPPVSAPWPSTPS----STAAKKVPAS*PSTTTTSNPLPASASSPTSSSSPSSPASPAP 357
Query: 534 IPQPPSSSPKVSPSASP-TIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQ 592
P S++P P A+P + S+P LP P ++S + P P
Sbjct: 358 RPLATSATP---PPANPCSKTSNPSWPTTASLPSPSLTTATSST-------TAPSEPDSN 507
Query: 593 KVSAPNNLV-----ISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESS 641
+AP++ +SS +SPL + + S++ S T +SS SS
Sbjct: 508 TTTAPSSTPPPTPRLSSTSSPLQNTGHSSSESWTPASISKAPTPSYSSRRTSSS 669
>TC216574 similar to UP|RK4_SPIOL (O49937) 50S ribosomal protein L4,
chloroplast precursor (R-protein L4), partial (74%)
Length = 1364
Score = 45.1 bits (105), Expect = 1e-04
Identities = 38/136 (27%), Positives = 58/136 (41%)
Frame = +1
Query: 532 TPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRG 591
+P PPSSS + S I+S+P + P+ P+ FP+ LL P
Sbjct: 106 SPSHHPPSSSLHTN-STHTIILSNPTAFPFYP-PKNPSPFPARPPLL---------FPSS 252
Query: 592 QKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIA 651
+ + S++A PLP PP+ + + S P+S A S E S + S
Sbjct: 253 TSPARGSASPSSTSAPPLPTPPEPSSTAPSSPTSRTSAAAPPPPSPAERSEAADGSPSRR 432
Query: 652 SPPLTPIALSNSRPSS 667
P+ P A +RPS+
Sbjct: 433 KKPVAPAAAPTARPSA 480
Score = 30.0 bits (66), Expect = 3.4
Identities = 35/133 (26%), Positives = 48/133 (35%), Gaps = 11/133 (8%)
Frame = +1
Query: 503 FSGPLTSNPMPTRPVSV---------DSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIV 553
F P +P P RP + S S P P P PSS+ SP+ S T
Sbjct: 190 FYPPKNPSPFPARPPLLFPSSTSPARGSASPSSTSAPPLPTPPEPSSTAPSSPT-SRTSA 366
Query: 554 SSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPN--NLVISSAASPLPI 611
++P E PS + P+ P + P+ SSA SP
Sbjct: 367 AAPPPPSPAERSEAADGSPSRRK--------KPVAPAAAPTARPSAPAEASSSARSPATG 522
Query: 612 PPQAMARSFSIPS 624
P ++ AR + PS
Sbjct: 523 PSRSTARRSASPS 561
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 45.1 bits (105), Expect = 1e-04
Identities = 44/138 (31%), Positives = 54/138 (38%), Gaps = 2/138 (1%)
Frame = +3
Query: 532 TPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRG 591
TP P PPS+ P + SP+ SSP S P P PS S S P P
Sbjct: 228 TPAPSPPSTMPPSATPPSPSATSSPSAS-----PTAPPTSPSPS------PPSSPSPPSA 374
Query: 592 QKVS--APNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISED 649
S AP+ S SP P PP P+S A + S S + +S S
Sbjct: 375 PSPSSTAPSPPSASPPTSPSPSPPS--------PASTASANSPASGSPTSRTSPTSPSPT 530
Query: 650 IASPPLTPIALSNSRPSS 667
S P P + S + PSS
Sbjct: 531 STSKPPAPSSSSPA*PSS 584
Score = 40.8 bits (94), Expect = 0.002
Identities = 46/150 (30%), Positives = 60/150 (39%), Gaps = 16/150 (10%)
Frame = +3
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLP--TPIPQPPSS-SPKVSPSASPTIVS----SP 556
S P T P T P + + P P +P P PPSS SP +PS S T S SP
Sbjct: 240 SPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASP 419
Query: 557 KISELHELPRPPTNFPSNSRLLGL-VGYSGPLVPRGQKVSAPNNLVISSAA--------S 607
S P P + +NS G + P P S P SS A S
Sbjct: 420 PTSPSPSPPSPASTASANSPASGSPTSRTSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPS 599
Query: 608 PLPIPPQAMARSFSIPSSGARVTALHSSSA 637
P PI P + + S P+S ++ + +S A
Sbjct: 600 PTPISPDP-SPATSTPTSHTSISPITASKA 686
>TC218421 similar to GB|AAQ65192.1|34365761|BT010569 At3g08670 {Arabidopsis
thaliana;} , partial (46%)
Length = 1256
Score = 43.5 bits (101), Expect = 3e-04
Identities = 53/220 (24%), Positives = 90/220 (40%), Gaps = 13/220 (5%)
Frame = +1
Query: 407 KSSISSGSNNPKPSKM---QTNSNEATTNLWHSSPLEQKKHETIRDEFSS-PTVRNAQSV 462
+ S++ ++ K S++ Q+ +N + SS + + T ++SS + R++ S+
Sbjct: 190 RRSLTRSTSTSKTSRLAVSQSENNNPASRPARSSSVTRSSISTSHSQYSSYSSNRHSSSI 369
Query: 463 LKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSV 522
L S+++ ++ P + SS + R +S P RPVS S
Sbjct: 370 LNTSSASVSSYIRPSSPITRSSSSARPSTPSSRPTASR-------SSTPSKPRPVSTSST 528
Query: 523 QMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRP---------PTNFPS 573
+ P P PSS P + P + SP S L RP P+ PS
Sbjct: 529 AERNRPSSQGSRPSTPSSRPHI-----PANLHSPSASSTRSLSRPSTPTRRSSMPSLSPS 693
Query: 574 NSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPP 613
S +G + +G + G+ SAP + S+ SP PP
Sbjct: 694 PSPTIGSLTSAGRVSSNGRN-SAPASR--PSSPSPRVRPP 804
Score = 35.8 bits (81), Expect = 0.063
Identities = 69/317 (21%), Positives = 110/317 (33%), Gaps = 3/317 (0%)
Frame = +1
Query: 317 EQEQDVSTSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADN 376
E E + + L + T A + EN + R S V S S +
Sbjct: 157 EGESQTTLAPPRRSLTRSTSTSKTSRLAVSQSENNNPASRPARSSSVTRSSISTSHSQYS 336
Query: 377 KPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNP--KPSKMQTNSNEATTNLW 434
S+ + + S SSY+ P+ +SS S+ + P +P+ ++++ +
Sbjct: 337 SYSSNRHSSSILNTSSASVSSYIRPSSPITRSSSSARPSTPSSRPTASRSSTPSKPRPVS 516
Query: 435 HSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAY 494
SS E+ + + S+P+ R S S ++T L P SS +
Sbjct: 517 TSSTAERNRPSSQGSRPSTPSSRPHIPANLHSPSASSTRSLSRPSTPTRRSSMPSLSPSP 696
Query: 495 SKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVS 554
S I +G ++SN + P S +P S SP+V P P +
Sbjct: 697 SPTIGSLTSAGRVSSNGRNSAPAS-----------------RPSSPSPRVRPPPQPIVP- 822
Query: 555 SPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQ 614
P P P N R + P P S P + + + +S P
Sbjct: 823 ----------PDFPLETPPNLRT------TLPDRPVSAGRSRPGGVTMKTNSSETQASPV 954
Query: 615 AMARSFSIP-SSGARVT 630
M R S P S RVT
Sbjct: 955 TMPRRHSSPIVSRGRVT 1005
>TC235011 weakly similar to UP|Q8V7F2 (Q8V7F2) ORF1 (Fragment), partial (72%)
Length = 824
Score = 43.1 bits (100), Expect = 4e-04
Identities = 42/162 (25%), Positives = 59/162 (35%)
Frame = +2
Query: 414 SNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATT 473
+ +P+PS T +T SS + +SP R+ S + +TATT
Sbjct: 359 AGSPRPSSPWTRRPTSTPRRSTSSARPSR*SSLTATTSASPAFRSTPSASTAPS*STATT 538
Query: 474 RLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTP 533
PP ++ R + S P + PT P + S P
Sbjct: 539 PSTPP----------------ARATSRSSRSAPRPTAWPPTPPSPASRPRWSSTACSTCP 670
Query: 534 IPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNS 575
PP+SSP S SA+PT+ S P P P T S S
Sbjct: 671 PRSPPTSSPATSTSAAPTVRSRPA-----HWPAPSTRPSSTS 781
>TC215176 similar to GB|BAC42264.1|26450296|AK117608 GPI-anchored protein
{Arabidopsis thaliana;} , partial (79%)
Length = 1570
Score = 43.1 bits (100), Expect = 4e-04
Identities = 34/118 (28%), Positives = 51/118 (42%), Gaps = 2/118 (1%)
Frame = +1
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSP-SASPTIVSSPKISEL-HE 563
PLT+ P T ++ + P P PP+ P P S++PT S+P S
Sbjct: 469 PLTTGPPTTSATALSPITACIWP------PSPPAIEPSTPPRSSAPTTSSAPTASSTASS 630
Query: 564 LPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFS 621
+ P + S S P PR ++ S P V++++ SPLP PP + S S
Sbjct: 631 VSSSPDPYRRTSTAAATSPPSPPSFPRERRRSIP---VLTASRSPLPFPPARLRSSQS 795
>TC221561 homologue to UP|Q9ZPA0 (Q9ZPA0) Inosine monophosphate dehydrogenase,
complete
Length = 2001
Score = 42.4 bits (98), Expect = 7e-04
Identities = 36/140 (25%), Positives = 51/140 (35%), Gaps = 2/140 (1%)
Frame = -1
Query: 434 WHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSA 493
W +P + H+ + S P + S ++ P L D L S +
Sbjct: 1140 WTQTPFQPSTHQPQPESDSQPGT*SQHSPQSHQAQGKPSSHTSPSLTDKLSSPENYPTLQ 961
Query: 494 YSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVS--PSASPT 551
+S + +A + P P RP TP P PPS +P VS PS SP
Sbjct: 960 HSTQPSPNAPTSPYHLPSSPPRPPP*------------TPRPPPPSPAPPVSGNPSPSPP 817
Query: 552 IVSSPKISELHELPRPPTNF 571
S P H P+P +F
Sbjct: 816 PPSPPNQQPHHPSPKPQHHF 757
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.129 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,650,264
Number of Sequences: 63676
Number of extensions: 513139
Number of successful extensions: 9254
Number of sequences better than 10.0: 608
Number of HSP's better than 10.0 without gapping: 6493
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8008
length of query: 669
length of database: 12,639,632
effective HSP length: 104
effective length of query: 565
effective length of database: 6,017,328
effective search space: 3399790320
effective search space used: 3399790320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0117b.11


