

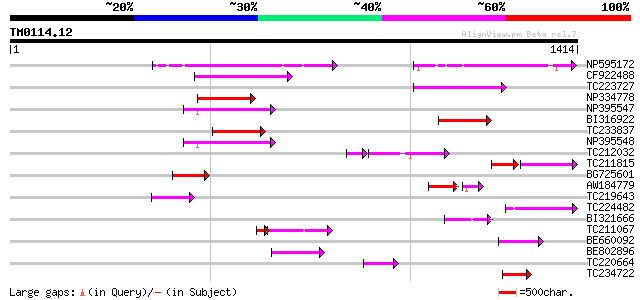
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0114.12
(1414 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 213 5e-55
CF922488 204 2e-52
TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 189 1e-47
NP334778 reverse transcriptase [Glycine max] 151 2e-36
NP395547 reverse transcriptase [Glycine max] 147 4e-35
BI316922 146 6e-35
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 139 1e-32
NP395548 reverse transcriptase [Glycine max] 134 4e-31
TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 105 7e-29
TC211815 75 5e-26
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 100 5e-21
AW184779 71 1e-20
TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotei... 84 6e-16
TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 83 1e-15
BI321666 81 3e-15
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 60 2e-11
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 67 4e-11
BE802896 65 2e-10
TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial... 65 2e-10
TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (... 58 3e-08
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 213 bits (542), Expect = 5e-55
Identities = 146/470 (31%), Positives = 236/470 (50%), Gaps = 9/470 (1%)
Frame = +1
Query: 357 QVQVKDKFLKIGSGLTTEQEDRLITLLGDNLDLFAWTINDVPGIDP--KVITHKLAIRPG 414
Q +V + LK L T + L LL +FA VP P + H + ++ G
Sbjct: 1627 QKEVPEDTLK---DLPTNIDPELAILLHTYAQVFA-----VPASLPPQREQDHAIPLKQG 1782
Query: 415 ATPV-IQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPT---WLANVVMVKKANGKWRM 470
+ PV ++P R +K + EK+I+ ++ + P+ + +++VKK +G WR
Sbjct: 1783 SGPVKVRPYRYPHTQKD-----QIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRF 1947
Query: 471 CTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMT 530
CTDY +LN + KDS+P+P VD+L+D G + S +D SGY+QI++ P D E TAF T
Sbjct: 1948 CTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRT 2127
Query: 531 NQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKE 590
+ +Y + MPFGL NA AT+Q LM+KIF + + + V+ DD+++ SA DH L+
Sbjct: 2128 HHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLES 2307
Query: 591 AFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQR 650
L+ +Q+ KCSFG +LG ++ G+ + K +A+L+ +P +VK+++
Sbjct: 2308 VLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRG 2487
Query: 651 LTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTP 710
G RF+ + A P L+K+S F W E E AF KLK+ + PVLS P
Sbjct: 2488 FLGLTGYYRRFIKSYANIAGPLTDLLQKDS-FLWNNEAEAAFVKLKKAMTEAPVLSLPDF 2664
Query: 711 GVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARR 770
P +L + V VL G+ I + S L + + LAI + +
Sbjct: 2665 SQPFILETDASGIGVGAVL----GQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSK 2832
Query: 771 LRPYFQSFQVKVKTDV-PLRQVLQKPDLSGRLVSWSVELSEYD--IQYEP 817
R Y + ++TD L+ ++ + + +W + YD I+Y+P
Sbjct: 2833 FRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKP 2982
Score = 119 bits (297), Expect = 1e-26
Identities = 110/428 (25%), Positives = 184/428 (42%), Gaps = 21/428 (4%)
Frame = +1
Query: 1007 LPWMESIKTFLE--------NPPKEDDLNTRTKRREASFYTLVDGELYRRG-IMSPMLKC 1057
L W E FLE +P + + T + +AS YT+ +G LY + ++ P +
Sbjct: 3028 LAWSEPHSIFLEELRARLISDPHLKQLMETYKQGADASHYTVREGLLYWKDRVVIPAEEE 3207
Query: 1058 VDTKDALGIMAEVHEGVCSSHIG-GRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFS 1116
+ K I+ E H H G R+LA ++A FYWP M++D Y++KC CQ
Sbjct: 3208 IVNK----ILQEYHSSPIGGHAGITRTLAR--LKAQFYWPKMQEDVKAYIQKCLICQQAK 3369
Query: 1117 DLHKAPPEELTTMMAPWPFAMW---GTDILGPFPVAKAQMKYIIVAVDYFTKWIEAEAV- 1172
+ P L + P P +W D + P + + I+V +D TK+ +
Sbjct: 3370 SNNTLPAGLLQPL--PIPQQVWEDVAMDFITGLPNSFG-LSVIMVVIDRLTKYAHFIPLK 3540
Query: 1173 ATITAAKVRNFLWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQT 1232
A + V IV G+P ++V D FTS+ + G + +S HPQ+
Sbjct: 3541 ADYNSKVVAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQS 3720
Query: 1233 NGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAML 1292
+GQ+E NK + L+ E W + LP + YNT S TP+R YG +
Sbjct: 3721 DGQSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPT 3900
Query: 1293 SVEIENQSWRVARFNENDNGENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRK 1352
A E + + + + + + ++ +A K ++ F
Sbjct: 3901 LTRQACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSF-------- 4056
Query: 1353 MRVGDLVL*K-------NTIPDKHNKLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTW 1405
++GD VL K + + K+ KLS + GP++++ +G AYKLE S ++ +
Sbjct: 4057 -QIGDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLELPSAARIHPVF 4233
Query: 1406 NASHLKQY 1413
+ S LK +
Sbjct: 4234 HVSQLKPF 4257
>CF922488
Length = 741
Score = 204 bits (519), Expect = 2e-52
Identities = 107/245 (43%), Positives = 147/245 (59%)
Frame = +3
Query: 461 VKKANGKWRMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHP 520
V K +GK MC DY LN PKD +PLP+++ LVD + S MD +SGYNQI + P
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 521 SDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSAR 580
D E T F+T +CYK M FGLKN GATYQR M +F + + +EVY+DDMIVKS
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 581 ASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMK 640
+H +L++ F +LR Y+++LNP KC F ++ K L F+ + RGIEV+ +K + ILEM
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 641 SPTSVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLA 700
P + K+VQ GR+ + RF+ P F L KN +W +C AF ++K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 701 TLPVL 705
VL
Sbjct: 723 NPHVL 737
>TC223727 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (9%)
Length = 843
Score = 189 bits (479), Expect = 1e-47
Identities = 92/236 (38%), Positives = 138/236 (57%), Gaps = 4/236 (1%)
Frame = +1
Query: 1008 PWMESIKTFL---ENPPKEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDAL 1064
PW IK ++ E P+ D + RT RR A+ + + LY+R L+CVD ++A
Sbjct: 136 PWYFDIKRYVISKEYLPEIADNDKRTLRRLAAGFFMSGSILYKRNHDMKPLRCVDAREAN 315
Query: 1065 GIMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPE 1124
++ EVHEG +H G ++A K++RAG+YW TM+ DC +V+KC KCQ F+D APP
Sbjct: 316 HMIEEVHEGSFGTHANGHAMARKILRAGYYWLTMESDCCVHVRKCHKCQAFADNVNAPPH 495
Query: 1125 ELTTMMAPWPFAMWGTDILGPF-PVAKAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNF 1183
L M +PWPF+MWG D++G P A ++I+VA+DYFTKW+EA + + V F
Sbjct: 496 PLNVMSSPWPFSMWGIDVIGAIEPKASNGHRFILVAIDYFTKWVEAASYTDVMRGVVVRF 675
Query: 1184 LWQRIVCRFGVPMALVMDNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESA 1239
+ + I+CR+G+P ++ DNGT + + E C E I+ + P+ N E A
Sbjct: 676 IKKEIICRYGLPRKIITDNGTNLNNKMMGEICEEFKIQHHNPTPYRPKMN*AVEVA 843
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 151 bits (381), Expect = 2e-36
Identities = 70/143 (48%), Positives = 92/143 (63%)
Frame = +3
Query: 469 RMCTDYTSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAF 528
RMC DY LN+ PKD++PLP++D L+ + L S MD +SGYNQI M P D E T F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 529 MTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDL 588
+T +CYK M FGLKN GATY R M +F + + +E YVD+MI KS +H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 589 KEAFDQLRTYQMKLNPEKCSFGI 611
+ F QLR Y+++LNP KC FG+
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 147 bits (370), Expect = 4e-35
Identities = 84/248 (33%), Positives = 126/248 (49%), Gaps = 18/248 (7%)
Frame = +1
Query: 433 VQLETEKLIKARFIREVQYPTWLANVVMVKKANG------------------KWRMCTDY 474
V+ E KL++A I + +W++ V +V K G +WRMC DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 475 TSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQAN 534
LN+ KD YPLP +D+++ + +D YSGYNQI + P D+E TAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 535 YCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQ 594
+ Y+ MPFGL NA T+QR M IF V + +EV++DD A + +L++ +
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 595 LRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGR 654
+ LN EKC F +Q G LG ++ RGIEV +K I ++ P +VK + G
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 655 MAALSRFL 662
+ RF+
Sbjct: 721 VGFYRRFI 744
>BI316922
Length = 405
Score = 146 bits (369), Expect = 6e-35
Identities = 66/134 (49%), Positives = 92/134 (68%)
Frame = +3
Query: 1069 EVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTT 1128
E+H G+C H + + +V+R G+Y TM+K C EYVKKCE+C F ++ EEL
Sbjct: 3 EMHRGICGMHSKSQLMTTRVLRVGYY**TMRKYCTEYVKKCEEC*KFGNISHLLVEELHN 182
Query: 1129 MMAPWPFAMWGTDILGPFPVAKAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRI 1188
++APWPFA+ G DIL PFP++K Q+KY++V +D FTKWIE E +A I+ A VR F+ + I
Sbjct: 183 IVAPWPFAI*GVDILRPFPLSKRQVKYLLVGIDQFTKWIETEHIAIISIANVRKFV*RNI 362
Query: 1189 VCRFGVPMALVMDN 1202
VC FG+P L+ DN
Sbjct: 363 VC*FGIPNTLISDN 404
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 139 bits (349), Expect = 1e-32
Identities = 65/132 (49%), Positives = 92/132 (69%)
Frame = +2
Query: 505 SLMDAYSGYNQIMMHPSDEESTAFMTNQANYCYKTMPFGLKNAGATYQRLMDKIFSKQVG 564
S MD +SGYNQI M D E T F+T + Y+ M FGLKN GATYQR M +F +
Sbjct: 5 SFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMMH 184
Query: 565 RNMEVYVDDMIVKSARASDHGGDLKEAFDQLRTYQMKLNPEKCSFGIQGGKFLGFMLTSR 624
+ +EVYVDDMI KS ++H +L + F +L+ YQ+KLNP KC+FG++ GK LGF+++ +
Sbjct: 185 KEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQK 364
Query: 625 GIEVNPDKGRAI 636
GIE++P+K +A+
Sbjct: 365 GIEIDPEKVKAL 400
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 134 bits (336), Expect = 4e-31
Identities = 77/248 (31%), Positives = 127/248 (51%), Gaps = 18/248 (7%)
Frame = +1
Query: 433 VQLETEKLIKARFIREVQYPTWLANVVMVKKANG------------------KWRMCTDY 474
V+ E KL++ I + W++ V++V K G W++C DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 475 TSLNKVCPKDSYPLPNVDKLVDGASGNELLSLMDAYSGYNQIMMHPSDEESTAFMTNQAN 534
LN+ KD +PLP +D++++ +G+ +DAY GYNQI++ P D+E AF
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 535 YCYKTMPFGLKNAGATYQRLMDKIFSKQVGRNMEVYVDDMIVKSARASDHGGDLKEAFDQ 594
+ Y+ +PFGL NA T+Q M IF+ V +++EV++DD V L+ +
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 595 LRTYQMKLNPEKCSFGIQGGKFLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEVQRLTGR 654
+ LN EKC F ++ G LG +++RGIEV+ K I ++ P++VK ++ G+
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 655 MAALSRFL 662
RF+
Sbjct: 721 ARFYRRFI 744
>TC212032 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (3%)
Length = 803
Score = 105 bits (263), Expect(2) = 7e-29
Identities = 66/210 (31%), Positives = 109/210 (51%), Gaps = 10/210 (4%)
Frame = +2
Query: 896 LRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRLMMEVKEIKIEHVPR 955
++ AI+ V+ L + GDS LV+ Q++GE + +DP L Y ++ L EI HV
Sbjct: 206 VQAAIDSNVKLLKVYGDSALVIHQLRGECETRDPNLIPYQAYIKELAGFFDEISFHHVA* 385
Query: 956 GQNERADVLAKLASTGRLGNYQTVIQETLPRPSIDLVEIKLK-------VVKSVNEGELP 1008
+N+ AD LA L S +L P + +E + + +V+ +G+ P
Sbjct: 386 EENQMADALATLVSMFQL----------TPHGDLPYIEFRCRGRPAHCCLVEEERDGK-P 532
Query: 1009 WMESIKTFLEN---PPKEDDLNTRTKRREASFYTLVDGELYRRGIMSPMLKCVDTKDALG 1065
W IK ++E+ P + D + R KRR A+ + + LY+R +L CV+ K+
Sbjct: 533 WYFDIKRYVESKEYPLEASDNDKRRKRRLAAGFFMSGSILYKRNHDMVLLHCVNGKEVEN 712
Query: 1066 IMAEVHEGVCSSHIGGRSLAVKVIRAGFYW 1095
++ EVHEG +H G ++A K++RAG+YW
Sbjct: 713 MLGEVHEGSFGTHSNGHAMARKILRAGYYW 802
Score = 41.6 bits (96), Expect(2) = 7e-29
Identities = 18/53 (33%), Positives = 30/53 (55%)
Frame = +1
Query: 839 EKTQGEWVLSVDGSSNNTGSGAGITIESPDKMIIEQSLKFEFKASNNQSEYEA 891
++ + +W++ D +SN G G G + SPD I + + F +NN +EYEA
Sbjct: 34 DEDRDKWIVWFDRASNVLGHGVGAILVSPDNQCIPFTTRLGFDCTNNMAEYEA 192
>TC211815
Length = 704
Score = 75.1 bits (183), Expect(2) = 5e-26
Identities = 37/67 (55%), Positives = 46/67 (68%)
Frame = +3
Query: 1201 DNGTQFTSSVTREFCAEMGIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAE 1260
DNG QFT+ EF + + I+ R SV+HPQTN +AE+ANKVIL LKK LD A G W E
Sbjct: 3 DNGLQFTNRKLNEFPSGLNIKHRVTSVKHPQTNRRAEAANKVILGDLKKLLDGANGRWVE 182
Query: 1261 ELPGVLW 1267
+L +LW
Sbjct: 183 DLVEILW 203
Score = 62.8 bits (151), Expect(2) = 5e-26
Identities = 46/143 (32%), Positives = 74/143 (51%), Gaps = 2/143 (1%)
Frame = +2
Query: 1274 QSSTKETPYRLTYGTDAMLSVEIENQSWRVARFNENDNGENLIANLIMLPEEQREAHIRN 1333
QS+T ETP+ L YG ML +E+ F E N E L +L ++ + + + I
Sbjct: 230 QSTTHETPF*LIYGISVMLPIEVGEVFL*RHYFAEV*NKEALQIDLDLIKQVREDTVIMT 409
Query: 1334 EAGKVKVARKFSTKVVPRKMRVGDLV--L*KNTIPDKHNKLSPNWGGPYRIIGDVGGEAY 1391
A K ++ R F++K+ G + + ++ +K + N GP++I + AY
Sbjct: 410 *AFKQRMTRCFNSKLPSTV*GRGPSMEGIQRSLEVLVRSKFTTN*EGPFKIRHNSKNGAY 589
Query: 1392 KLEQLSGQKVPRTWNASHLKQYF 1414
KLE+LSG+ V R WN+ HLK Y+
Sbjct: 590 KLEELSGKVVLRIWNSMHLKVYY 658
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 100 bits (249), Expect = 5e-21
Identities = 50/91 (54%), Positives = 61/91 (66%)
Frame = -3
Query: 407 HKLAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMVKKANG 466
HKLAI V Q R++ EE+ + V E KL A FIR++ Y T L +VVMVKK NG
Sbjct: 277 HKLAICNDVKLVTQRKRKIREERCQTV*QEVVKLAIASFIRDINYST*LFSVVMVKKPNG 98
Query: 467 KWRMCTDYTSLNKVCPKDSYPLPNVDKLVDG 497
KWR+CTDY LN CPKD+YPLPN+D + DG
Sbjct: 97 KWRICTDYIDLN*ACPKDAYPLPNIDHMTDG 5
>AW184779
Length = 432
Score = 71.2 bits (173), Expect(2) = 1e-20
Identities = 33/74 (44%), Positives = 48/74 (64%)
Frame = +1
Query: 1045 LYRRGIMSPMLKCVDTKDALGIMAEVHEGVCSSHIGGRSLAVKVIRAGFYWPTMKKDCLE 1104
LY+R +L+CVD ++A ++ EVHEG +H ++A K++R G+YW TM+ DC
Sbjct: 16 LYKRNHDMVLLRCVDAREAEQMLVEVHEGSFGTHANIHAMAQKILRVGYYWLTMESDCCI 195
Query: 1105 YVKKCEKCQVFSDL 1118
+V KC KCQ SDL
Sbjct: 196 HVWKCHKCQ--SDL 231
Score = 48.1 bits (113), Expect(2) = 1e-20
Identities = 24/62 (38%), Positives = 36/62 (57%), Gaps = 10/62 (16%)
Frame = +3
Query: 1129 MMAPWPFA---------MWGTDILGPF-PVAKAQMKYIIVAVDYFTKWIEAEAVATITAA 1178
M+ P P+ MWG D++G P A +I+VA+DYFTKW+EA + A++T +
Sbjct: 240 MLTPHPYL*MSWQHLGHMWGIDVIGAIEPKASNGHHFILVAIDYFTKWVEAVSYASVTRS 419
Query: 1179 KV 1180
V
Sbjct: 420 VV 425
>TC219643 weakly similar to UP|Q6WAY7 (Q6WAY7) Gag/pol polyprotein (Fragment),
partial (8%)
Length = 1320
Score = 83.6 bits (205), Expect = 6e-16
Identities = 41/113 (36%), Positives = 66/113 (58%), Gaps = 4/113 (3%)
Frame = +3
Query: 353 EETKQVQVKD----KFLKIGSGLTTEQEDRLITLLGDNLDLFAWTINDVPGIDPKVITHK 408
EET+ V + + +KIG+G+T + LI LL D D+FAW+ D+PG+ ++ H+
Sbjct: 978 EETELVDLGSGSGKREVKIGTGITAPIREELIILLRDYQDIFAWSYQDMPGLSSDIVQHR 1157
Query: 409 LAIRPGATPVIQPMRRMSEEKHKAVQLETEKLIKARFIREVQYPTWLANVVMV 461
L + P +PV Q +RRM E ++ E +K A F+ +YP W+AN+V +
Sbjct: 1158 LPLNPECSPVKQKLRRMKPETSLKIKEEVKK*FDAGFLAVARYPKWVANIVPI 1316
>TC224482 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 669
Score = 82.8 bits (203), Expect = 1e-15
Identities = 56/183 (30%), Positives = 92/183 (49%), Gaps = 5/183 (2%)
Frame = +1
Query: 1237 ESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYGTDAMLSVEI 1296
E+ANK I K ++K K W E LP L Y T+ ++ST TP+ L YG +A+L E+
Sbjct: 1 EAANKNIKKIIQKMTVSYKD-WHEMLPFALHGYRTSVRTSTGATPFSLVYGMEAVLPFEV 177
Query: 1297 ENQSWRV---ARFNENDNGENLIANLIMLPEEQREAHIRNEAGKVKVARKFSTKVVPRKM 1353
E S R+ + E++ + L ++ ++ A + ++ F KV RK
Sbjct: 178 EVPSLRILAESGLKESEWAQTRYDQLNLIEGKRLTAMSHGRLYQQRMKSAFDKKVCLRKF 357
Query: 1354 RVGDLVL*K--NTIPDKHNKLSPNWGGPYRIIGDVGGEAYKLEQLSGQKVPRTWNASHLK 1411
GDLVL K + + D K +PN+ GP+ + G A L + G+++P N+ +K
Sbjct: 358 HEGDLVLKKMSHAVKDHRGKWAPNYEGPFVVKRAFSGGALVLTNMDGEELPSPMNSDVVK 537
Query: 1412 QYF 1414
+Y+
Sbjct: 538 RYY 546
>BI321666
Length = 430
Score = 81.3 bits (199), Expect = 3e-15
Identities = 45/120 (37%), Positives = 67/120 (55%)
Frame = +2
Query: 1084 LAVKVIRAGFYWPTMKKDCLEYVKKCEKCQVFSDLHKAPPEELTTMMAPWPFAMWGTDIL 1143
++ V+++ FY P++ KD + + C KCQ + K L T++ F WG D +
Sbjct: 2 ISTNVLQSRFYLPSIFKDAYVHAQSCNKCQRTRSVSKRNELPLHTILEVEIFDYWGIDFV 181
Query: 1144 GPFPVAKAQMKYIIVAVDYFTKWIEAEAVATITAAKVRNFLWQRIVCRFGVPMALVMDNG 1203
GPFP + + +YI+V VDY +KW+EA A A V FL ++I R GVP L+ DNG
Sbjct: 182 GPFPPSFSN-EYILVVVDYVSKWVEAVACQKSDAKIVIKFLKKQIFSRLGVPWVLI-DNG 355
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 60.5 bits (145), Expect(2) = 2e-11
Identities = 47/162 (29%), Positives = 70/162 (43%), Gaps = 1/162 (0%)
Frame = +1
Query: 644 SVKEVQRLTGRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLP 703
SV +++ G + RF+P A+P +KKN F W E+ EQAF LKE L P
Sbjct: 109 SVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAP 288
Query: 704 VLSKPTPGVPLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALA 763
VL+ P L + V VLLQ + YF S L A L Y +K A
Sbjct: 289 VLALPDFSKTFELECDASGVGVRAVLLQ---GGHPIAYF-SEKLHSATLNYPTYDKELYA 456
Query: 764 ILKTARRLRPYFQSFQVKVKTD-VPLRQVLQKPDLSGRLVSW 804
+++ + + + + +D L+ + K L+ R W
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKW 582
Score = 27.7 bits (60), Expect(2) = 2e-11
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = +2
Query: 616 FLGFMLTSRGIEVNPDKGRAILEMKSPTSVKEV 648
F GF++ G++++P+K +AI E P V E+
Sbjct: 23 FSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI 121
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 67.4 bits (163), Expect = 4e-11
Identities = 38/115 (33%), Positives = 64/115 (55%), Gaps = 3/115 (2%)
Frame = -3
Query: 1219 GIEMRFASVEHPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTK 1278
G+ R ++ HPQTNGQAE +N+ I + L+K + ++ W+ L LWA+ T ++
Sbjct: 352 GVVHRVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTAYKAPIG 173
Query: 1279 ETPYRLTYGTDAMLSVEIENQSW---RVARFNENDNGENLIANLIMLPEEQREAH 1330
+PYR+ +G L VEIE++++ + F+ + GE L L E + EA+
Sbjct: 172 MSPYRVVFGKACHLPVEIEHKAYWAVKTCNFSMDQAGEERKLQLSELDEIRLEAY 8
>BE802896
Length = 416
Score = 65.1 bits (157), Expect = 2e-10
Identities = 40/133 (30%), Positives = 60/133 (45%)
Frame = -2
Query: 653 GRMAALSRFLPMAGDKAAPFFTCLKKNSKFQWTEECEQAFTKLKETLATLPVLSKPTPGV 712
G RF+ A P L+K +F + ++C+ AF LK L T P++ P
Sbjct: 406 GHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPDWTA 227
Query: 713 PLVLYLAVTDKAVSTVLLQEEGKKQKVIYFVSHTLQGAELRYQKIEKAALAILKTARRLR 772
P L ++ A+ VL Q+ K +VIY+ S TL A+ Y EK LAI+ +
Sbjct: 226 PFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALEKFH 47
Query: 773 PYFQSFQVKVKTD 785
Y ++ V D
Sbjct: 46 SYLLGTRIIVYID 8
>TC220664 weakly similar to UP|Q84TE0 (Q84TE0) At5g51080, partial (31%)
Length = 724
Score = 65.1 bits (157), Expect = 2e-10
Identities = 34/87 (39%), Positives = 51/87 (58%)
Frame = +1
Query: 882 ASNNQSEYEALIAGLRLAIELGVQKLFIKGDSQLVVKQVKGEYQVKDPQLSKYLEVVRRL 941
A+NN +EY A+I G++ A++ G + I+GDS+LV Q+ G ++VK+ LS V + L
Sbjct: 61 ATNNAAEYRAMILGMKYALKKGFTGIRIQGDSKLVCMQIDGSWKVKNENLSTLYNVAKEL 240
Query: 942 MMEVKEIKIEHVPRGQNERADVLAKLA 968
+ +I HV R N AD A LA
Sbjct: 241 KDKFSSFQISHVLRNFNSDADAQANLA 321
>TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (32%)
Length = 482
Score = 57.8 bits (138), Expect = 3e-08
Identities = 28/73 (38%), Positives = 45/73 (61%)
Frame = -3
Query: 1229 HPQTNGQAESANKVILKGLKKKLDEAKGLWAEELPGVLWAYNTTEQSSTKETPYRLTYGT 1288
HPQTNGQAE +NK I + L+ + ++ WA +L WAY ++ +P++L YG
Sbjct: 231 HPQTNGQAEVSNKEIKRVLENIVVSSRKDWALKLDDAFWAYRIAFKTPIGLSPFQLVYGK 52
Query: 1289 DAMLSVEIENQSW 1301
LSVE+E++++
Sbjct: 51 ACHLSVELEHKAY 13
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,561,751
Number of Sequences: 63676
Number of extensions: 710145
Number of successful extensions: 3399
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 3336
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3390
length of query: 1414
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1305
effective length of database: 5,698,948
effective search space: 7437127140
effective search space used: 7437127140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0114.12


