

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.6
(545 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
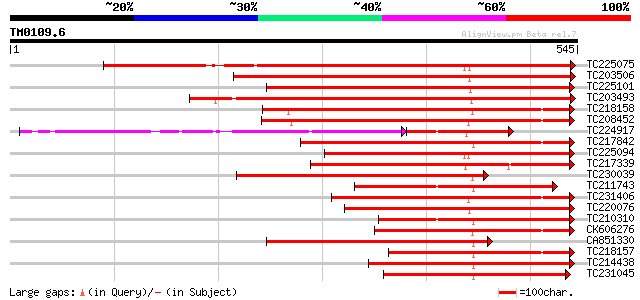
Score E
Sequences producing significant alignments: (bits) Value
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 440 e-124
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 388 e-108
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 379 e-105
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 377 e-105
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 350 7e-97
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 348 5e-96
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 231 5e-94
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 310 1e-84
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 308 3e-84
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 289 3e-78
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 281 4e-76
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 258 5e-69
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 251 4e-67
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 244 5e-65
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 244 7e-65
CK606276 243 1e-64
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 238 7e-63
TC218157 similar to UP|Q8GUF8 (Q8GUF8) At2g47550/T30B22.15, part... 218 7e-57
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 216 2e-56
TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase iso... 215 5e-56
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 440 bits (1131), Expect = e-124
Identities = 224/464 (48%), Positives = 307/464 (65%), Gaps = 10/464 (2%)
Frame = +1
Query: 91 LTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMRRIRAGDTSV-QYEGNLEAWLS 149
L++F + + R AI DC +L+D S L+W+L + + S +L WLS
Sbjct: 205 LSRFGSPLANFRLSTAIADCLDLLDLSSDVLSWALSASQNPKGKHNSTGNLSSDLRTWLS 384
Query: 150 AALSNQDTCLEGFEGTDRRLESYISGSLTQVTQLISNVLSLYTQLHALPFRPPRNNTHKT 209
AAL++ +TC+EGFEGT+ ++ +S + QV L+ +L+ LP
Sbjct: 385 AALAHPETCMEGFEGTNSIVKGLVSAGIGQVVSLVEQLLA-----QVLP-------AQDQ 528
Query: 210 SESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRR 269
++ +FP W+ +++LL++ D VALDGSG+Y I +AV AAP +S +R
Sbjct: 529 FDAASSKGQFPSWIKPKERKLLQAIA--VTPDVTVALDGSGNYAKIMDAVLAAPDYSMKR 702
Query: 270 YVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGF 329
+VI VKKG+Y ENV++K+K NIM++G G+ T+I+ NR+ + GWTTFR+AT AVSG+GF
Sbjct: 703 FVILVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFAVSGRGF 882
Query: 330 IARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTI 389
IARD+SF+NTAGP HQAVALR DSD S F+RC I G+QD+LY H++RQF+R+C I GT+
Sbjct: 883 IARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTISGTV 1062
Query: 390 DFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQ------DSFIL--- 440
D+IFG+ AV QNC + + LP QK T+TA GRK P++ TGF+ Q DS ++
Sbjct: 1063DYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADSDLIPSV 1242
Query: 441 ATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLA 500
T TYLGRPWK YSRTV++ +YMS ++ GWLEW G+FAL TL+Y EY N G GA +A
Sbjct: 1243GTAQTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWNGNFALDTLYYAEYMNTGAGAGVA 1422
Query: 501 GRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGLT 544
RVKWPGYH + D++ AS FTV +FI G+ WLP TGV FTAGLT
Sbjct: 1423NRVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGLT 1554
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 388 bits (997), Expect = e-108
Identities = 183/339 (53%), Positives = 236/339 (68%), Gaps = 10/339 (2%)
Frame = +2
Query: 216 DSEFPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVK 275
+ +FP W+ + D+ LL +AD V+ G+G +TI EA+ AP HS RR++IYV+
Sbjct: 2 EGKFPGWLNKRDRRLLSLPLSAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYVR 181
Query: 276 KGLYREN-VDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDM 334
G Y EN + + RK TN+M +GDG G+T+IT +N + G TTF +A+ A SG GFIARD+
Sbjct: 182 AGRYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARDI 361
Query: 335 SFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFG 394
+F N AGP HQAVALRV +D + YRC+I G+QD+ Y HS RQF+REC IYGT+DFIFG
Sbjct: 362 TFENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIFG 541
Query: 395 NGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFIL---------ATQPT 445
N A V Q C IY R P+ QK T+TAQ RK P+Q+TG ++ + IL + PT
Sbjct: 542 NAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFPT 721
Query: 446 YLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKW 505
YLGRPWKQYSRTVY+ +YM + PRGWLEW GDFAL TL+YGEY NYGPGA++ RV+W
Sbjct: 722 YLGRPWKQYSRTVYLVSYMGDHIHPRGWLEWNGDFALNTLYYGEYMNYGPGAAVGQRVQW 901
Query: 506 PGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGLT 544
PGY ++K A+ FTV +FI G +WLP TGV F AGL+
Sbjct: 902 PGYRVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGLS 1018
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 379 bits (974), Expect = e-105
Identities = 179/308 (58%), Positives = 233/308 (75%), Gaps = 12/308 (3%)
Frame = +3
Query: 248 GSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSN 307
G +Y + +AV AAP++S +RYVI++K+G+Y ENV++K+K N+M+VGDG+ TII+ N
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 308 RNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGF 367
R+F+ GWTTFR+AT AVSG+GFIARD++F+NTAGP HQAVALR DSD S F+RC I G+
Sbjct: 387 RSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGY 566
Query: 368 QDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPH 427
QD+LY H++RQFYREC+I GT+DFIFG+ A+ QNC I + LP QK T+TA GRK+P
Sbjct: 567 QDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNPD 746
Query: 428 QSTGFTIQDSFILA------------TQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLE 475
+ TGF+IQ I A + TYLGRPWK YSRT+++ +Y+S +++P GWLE
Sbjct: 747 EPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWLE 926
Query: 476 WFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGT 535
W GDFAL TL+Y EY NYGPGA +A RVKW GYH+M D++ AS FTV +FI G+ LP T
Sbjct: 927 WNGDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCLPST 1106
Query: 536 GVKFTAGL 543
GV FTAGL
Sbjct: 1107GVTFTAGL 1130
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 377 bits (969), Expect = e-105
Identities = 191/385 (49%), Positives = 252/385 (64%), Gaps = 14/385 (3%)
Frame = +1
Query: 174 SGSLTQVTQLISNVLSLYTQLHA---LPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQEL 230
S +L +++L+SN L++++ A P +N + D FP W+ D+ L
Sbjct: 4 SNNLKDLSELVSNCLAIFSGAGAGDDFAGVPIQNRRRLMAMRED---NFPTWLNGRDRRL 174
Query: 231 LRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRE-NVDMKRKM 289
L + +AD VV+ DG+G +TI EA+ P +S+RR +IY++ G Y E N+ + RK
Sbjct: 175 LSLPLSQIQADIVVSKDGNGTVKTIAEAIKKVPEYSSRRIIIYIRAGRYEEDNLKLGRKK 354
Query: 290 TNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVA 349
TN+M +GDG G+T+IT RN+ Q TTF TA+ A SG GFIA+DM+F N AGP HQAVA
Sbjct: 355 TNVMFIGDGKGKTVITGGRNYYQNLTTFHTASFAASGSGFIAKDMTFENYAGPGRHQAVA 534
Query: 350 LRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRA 409
LRV +D + YRC+I G+QDT+Y HS RQFYREC+IYGT+DFIFGN A V QNC ++ R
Sbjct: 535 LRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCTLWARK 714
Query: 410 PLPLQKVTVTAQGRKSPHQSTGFTIQDSFILAT---------QPTYLGRPWKQYSRTVYI 460
P+ QK T+TAQ RK P+Q+TG +I + I+AT PTYLGRPWK Y+RTV++
Sbjct: 715 PMAQQKNTITAQNRKDPNQNTGISIHNCRIMATPDLEASKGSYPTYLGRPWKLYARTVFM 894
Query: 461 NTYMSGMVQPRGWLEW-FGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASY 519
+Y+ V PRGWLEW FAL T +YGEY NYGPG++L RV W GY + AS
Sbjct: 895 LSYIGDHVHPRGWLEWNTSSFALDTCYYGEYMNYGPGSALGQRVNWAGYRAINSTVEASR 1074
Query: 520 FTVQRFIHGDSWLPGTGVKFTAGLT 544
FTV +FI G SWLP TGV F AGL+
Sbjct: 1075FTVGQFISGSSWLPSTGVAFIAGLS 1149
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 350 bits (899), Expect = 7e-97
Identities = 170/311 (54%), Positives = 217/311 (69%), Gaps = 11/311 (3%)
Frame = +3
Query: 244 VALDGSGHYRTITEAVNAAPSHS---NRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIG 300
V+ DGSG++ TI +A+ AAP+ S + ++IYV G+Y ENV + +K T +M+VGDGI
Sbjct: 159 VSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEENVSIDKKKTYLMMVGDGIN 338
Query: 301 QTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFY 360
+TIIT NR+ + GWTTF +AT+AV G+GF+ +M+ RNTAG V HQAVALR +D S FY
Sbjct: 339 KTIITGNRSVVDGWTTFSSATLAVVGQGFVGVNMTIRNTAGAVKHQAVALRSGADLSTFY 518
Query: 361 RCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTA 420
CS EG+QDTLY HSLRQFY EC+IYGT+DFIFGN V QNCK+Y R P+ Q +TA
Sbjct: 519 SCSFEGYQDTLYVHSLRQFYSECDIYGTVDFIFGNAKVVFQNCKMYPRLPMSGQFNAITA 698
Query: 421 QGRKSPHQSTGFTIQDSFILATQ--------PTYLGRPWKQYSRTVYINTYMSGMVQPRG 472
QGR P+Q TG +I + I A TYLGRPWK+YSRTVY+ T M ++ +G
Sbjct: 699 QGRTDPNQDTGISIHNCTIRAADDLAASNGVATYLGRPWKEYSRTVYMQTVMDSVIHAKG 878
Query: 473 WLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWL 532
W EW GDFAL+TL+Y EY N GPG+ RV WPGYH++ +A A+ FTV F+ GD WL
Sbjct: 879 WREWDGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYHVI-NATDAANFTVSNFLLGDDWL 1055
Query: 533 PGTGVKFTAGL 543
P TGV +T L
Sbjct: 1056PQTGVSYTNNL 1088
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 348 bits (892), Expect = 5e-96
Identities = 169/313 (53%), Positives = 224/313 (70%), Gaps = 12/313 (3%)
Frame = +1
Query: 243 VVALDGSGHYRTITEAVNAAPSHSNRR---YVIYVKKGLYRENVDMKRKMTNIMLVGDGI 299
+V+ G +Y +I +A+ AAP+++ +++YV++GLY E V + ++ NI+LVGDGI
Sbjct: 295 IVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGLYEEYVVIPKEKKNILLVGDGI 474
Query: 300 GQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAF 359
+TIIT N + + GWTTF ++T AVSG+ FIA D++FRNTAGP HQAVA+R ++D S F
Sbjct: 475 NKTIITGNHSVIDGWTTFNSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAVRNNADLSTF 654
Query: 360 YRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVT 419
YRCS EG+QDTLY HSLRQFYRECEIYGT+DFIFGN A V Q CKIY R PLP QK VT
Sbjct: 655 YRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKPLPNQKNAVT 834
Query: 420 AQGRKSPHQSTGFTIQDSFI---------LATQPTYLGRPWKQYSRTVYINTYMSGMVQP 470
AQGR P+Q+TG +IQ+ I L + ++LGRPWK YSRTVY+ +Y+ ++QP
Sbjct: 835 AQGRTDPNQNTGISIQNCSIDAAPDLVADLNSTMSFLGRPWKVYSRTVYLQSYIGNVIQP 1014
Query: 471 RGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDS 530
GWLEW G L TL+YGE+ NYGPG++ + RV WPGY ++ +A A FTV F G++
Sbjct: 1015AGWLEWNGTVGLDTLFYGEFNNYGPGSNTSNRVTWPGYSLL-NATQAWNFTVLNFTLGNT 1191
Query: 531 WLPGTGVKFTAGL 543
WLP T + +T GL
Sbjct: 1192WLPDTDIPYTEGL 1230
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 231 bits (590), Expect(2) = 5e-94
Identities = 151/373 (40%), Positives = 202/373 (53%), Gaps = 1/373 (0%)
Frame = +2
Query: 10 LMMLLILPSSSKSFSDEIPSEIQTQDMQALIAQACMDIENQNSCLQNIHNELTKIGPPSP 69
LM LL+ P S I S +D+Q+ C C + N S
Sbjct: 134 LMTLLLAPFLFSS----IASSYSFKDIQSW----CNQTPYPQPCEYYLTNHAFNKPIKSK 289
Query: 70 TSVLSAALRTTLNEAQGAIDNLTKFSTFSVSNREQIAIGDCKELVDFSVSELAWSLGEMR 129
+ L +L+ L AQ + N + E+ A DC +L ++++ L ++
Sbjct: 290 SDFLKVSLQLALERAQRSELNTHALGPKCRNVHEKAAWADCLQLYEYTIQRLNKTINPNT 469
Query: 130 RIRAGDTSVQYEGNLEAWLSAALSNQDTCLEGFEGTDRRLESYISGSLTQ-VTQLISNVL 188
+ DT + WLS AL+N +TC GF + + Y+ ++ VT+L+SN L
Sbjct: 470 KCNQTDT--------QTWLSTALTNLETCKNGFY--ELGVPDYVLPLMSNNVTKLLSNTL 619
Query: 189 SLYTQLHALPFRPPRNNTHKTSESLDLDSEFPEWMTEGDQELLRSKPHRSRADAVVALDG 248
SL + ++PP FP W+ GD++LL+S S A+ VVA DG
Sbjct: 620 SLNKGPYQ--YKPP-----------SYKEGFPTWVKPGDRKLLQSSSVASNANVVVAKDG 760
Query: 249 SGHYRTITEAVNAAPSHSNRRYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITSNR 308
SG Y T+ AV+AAP S+ RYVIYVK G+Y E V++K NIMLVGDGIG+TIIT ++
Sbjct: 761 SGKYTTVKAAVDAAPKSSSGRYVIYVKSGVYNEQVEVKGN--NIMLVGDGIGKTIITGSK 934
Query: 309 NFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQ 368
+ G TTFR+ATVA G GFIA+D++FRNTAG NHQAVA R SD S FYRCS EGFQ
Sbjct: 935 SVGGGTTTFRSATVAAVGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYRCSFEGFQ 1114
Query: 369 DTLYAHSLRQFYR 381
DTLY HS RQFYR
Sbjct: 1115DTLYVHSERQFYR 1153
Score = 132 bits (331), Expect(2) = 5e-94
Identities = 61/110 (55%), Positives = 80/110 (72%), Gaps = 7/110 (6%)
Frame = +3
Query: 382 ECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDS---- 437
EC+IYGT+DFIFGN AAVLQNC IY R P P + +TVTAQGR P+Q+TG I +S
Sbjct: 1155 ECDIYGTVDFIFGNAAAVLQNCNIYARTP-PQRTITVTAQGRTDPNQNTGIIIHNSKVTG 1331
Query: 438 ---FILATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTT 484
F ++ +YLGRPW++YSRTV++ TY+ ++ P GW+EW G+FAL T
Sbjct: 1332 ASGFNPSSVKSYLGRPWQKYSRTVFMKTYLDSLINPAGWMEWDGNFALDT 1481
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 310 bits (794), Expect = 1e-84
Identities = 147/273 (53%), Positives = 192/273 (69%), Gaps = 9/273 (3%)
Frame = +3
Query: 280 RENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNT 339
+EN+D+KR + N+M+VGDG+G TI+T N N + G TTFR+AT AV G GFIARD++F NT
Sbjct: 6 KENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFENT 185
Query: 340 AGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAV 399
AGP HQAVALR +D S FYRCS G+QDTLY ++ RQFYR+C+IYGT+DFIFG+ AV
Sbjct: 186 AGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVAV 365
Query: 400 LQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQP---------TYLGRP 450
LQNC IY R P+ Q+ TVTAQGR P+++TG I + I A T+LGRP
Sbjct: 366 LQNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGRP 545
Query: 451 WKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHI 510
W++YSRTV + + + G++ P GW W G+FAL+TL+Y E+ N G GAS GRV W G+ +
Sbjct: 546 WQKYSRTVVMKSALDGLISPAGWFPWSGNFALSTLYYAEHANTGAGASTGGRVDWAGFRV 725
Query: 511 MKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
+ + A FTV F+ G SW+PG+GV F GL
Sbjct: 726 I-SSTEAVKFTVGNFLAGGSWIPGSGVPFDEGL 821
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 308 bits (790), Expect = 3e-84
Identities = 147/250 (58%), Positives = 187/250 (74%), Gaps = 9/250 (3%)
Frame = +3
Query: 303 IITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRC 362
II+ NR+ + GWTTFR+AT AVSG+GFIARD+SF+NTAGP HQAVALR D+D S F+RC
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 363 SIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQG 422
I G+QD+LY H++RQF+REC I GT+D+IFG+ AV QNC + + LP QK T+TA G
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 423 RKSPHQSTGFTIQ------DSFI---LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGW 473
RK P++ TGF+ Q DS + +++ +YLGRPWK YSRTV++ +YMS +++ GW
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 474 LEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLP 533
LEW G+FAL TL+YGEY N G GA LA RVKWPGYH D+ AS FTV +FI G+ WLP
Sbjct: 543 LEWNGNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLWLP 722
Query: 534 GTGVKFTAGL 543
TGV +TAGL
Sbjct: 723 STGVTYTAGL 752
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 289 bits (739), Expect = 3e-78
Identities = 148/266 (55%), Positives = 179/266 (66%), Gaps = 12/266 (4%)
Frame = +1
Query: 290 TNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVA 349
TN+MLVGDG T+IT N NF+ G TTF+TATVA G GFIA+D+ F+NTAGP HQAVA
Sbjct: 7 TNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQAVA 186
Query: 350 LRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRA 409
LRV +DQS RC I+ FQDTLYAHS RQFYR+ I GT+DFIFGN A V Q C + R
Sbjct: 187 LRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVARK 366
Query: 410 PLPLQKVTVTAQGRKSPHQSTGFTIQD---------SFILATQPTYLGRPWKQYSRTVYI 460
P+ Q VTAQGR+ P+Q+TG +IQ ++ + T+LGRPWK+YSRTV +
Sbjct: 367 PMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTVVM 546
Query: 461 NTYMSGMVQPRGWLEWFG---DFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATA 517
+ + + P GW EW DF L TL+YGEY N GPGA + RV WPGYHI+K AA A
Sbjct: 547 QSTLDSHIDPTGWAEWDAQSKDF-LQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAAEA 723
Query: 518 SYFTVQRFIHGDSWLPGTGVKFTAGL 543
S FTV + I G+ WL TGV F GL
Sbjct: 724 SKFTVAQLIQGNVWLKNTGVNFIEGL 801
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 281 bits (720), Expect = 4e-76
Identities = 134/251 (53%), Positives = 182/251 (72%), Gaps = 9/251 (3%)
Frame = +1
Query: 219 FPEWMTEGDQELLRSKPHRSRADAVVALDGSGHYRTITEAVNAAPSHSNRRYVIYVKKGL 278
FP W++ D++LL++ + + + +VA DG+G++ TI EAV AP+ S R+VI++K G
Sbjct: 160 FPTWLSTKDRKLLQAAVNETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIHIKAGA 339
Query: 279 YRENVDMKRKMTNIMLVGDGIGQTIITSNRNFMQGWTTFRTATVAVSGKGFIARDMSFRN 338
Y ENV++ RK TN+M VGDGIG+T++ ++RN + GWTTF++ATVAV G GFIA+ ++F N
Sbjct: 340 YFENVEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKGITFEN 519
Query: 339 TAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAA 398
+AGP HQAVALR SD SAFY+CS +QDTLY HSLRQFYR+C++YGT+DFIFGN A
Sbjct: 520 SAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIFGNAAT 699
Query: 399 VLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ---------PTYLGR 449
VLQNC +Y R P Q+ TAQGR+ P+Q+TG +I + + A YLGR
Sbjct: 700 VLQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGR 879
Query: 450 PWKQYSRTVYI 460
PWK+YSRTVY+
Sbjct: 880 PWKKYSRTVYL 912
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 258 bits (659), Expect = 5e-69
Identities = 122/200 (61%), Positives = 149/200 (74%), Gaps = 5/200 (2%)
Frame = +2
Query: 332 RDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDF 391
+D++FRNTAG NHQAVALR SD S FYRCS EG+QDTLY +S RQFYREC+IYGT+DF
Sbjct: 2 QDITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDF 181
Query: 392 IFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQP-----TY 446
IFGN A V QNC IY R P P + T+TAQGR P+Q+TG +I +S + A TY
Sbjct: 182 IFGNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKVTAASDLMGVRTY 358
Query: 447 LGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWP 506
LGRPW+QYSRTV++ TY+ ++ P GWLEW G+FAL+TL+YGEY N GPG+S A RV W
Sbjct: 359 LGRPWQQYSRTVFMKTYLDSLINPEGWLEWSGNFALSTLYYGEYMNTGPGSSTANRVNWL 538
Query: 507 GYHIMKDAATASYFTVQRFI 526
GYH++ A+ AS FTV FI
Sbjct: 539 GYHVITSASEASKFTVGNFI 598
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 251 bits (642), Expect = 4e-67
Identities = 122/239 (51%), Positives = 158/239 (66%), Gaps = 5/239 (2%)
Frame = +2
Query: 310 FMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQD 369
F+ G TF TAT AV G+ FIARDM FRNTAGP QAVAL +DQ+ +YRC I+ FQD
Sbjct: 2 FVDGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQD 181
Query: 370 TLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQS 429
+LYAHS RQFYREC IYGT+DFIFGN A VLQNC I R P+ Q+ +TAQG+ P+ +
Sbjct: 182 SLYAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMN 361
Query: 430 TGFTIQDSFI-----LATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDFALTT 484
TG +IQ+ I L++ TYLGRPWK YS V++ + M + P GWL W G+ A T
Sbjct: 362 TGISIQNCNITPFGDLSSVKTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNSAPDT 541
Query: 485 LWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
++Y E++N GPGAS RV W G ++ AS FTV+ F+ G+ W+ +G F + +
Sbjct: 542 IFYAEFQNVGPGASTKNRVNWKGLRVI-TRKQASMFTVKAFLSGERWITASGAPFKSSI 715
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 244 bits (624), Expect = 5e-65
Identities = 123/227 (54%), Positives = 150/227 (65%), Gaps = 6/227 (2%)
Frame = +2
Query: 323 AVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRE 382
AV GKGFIA+D+ F N AG HQAVA R SD+S F+RCS GFQDTLYAHS RQFYR+
Sbjct: 80 AVKGKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRCSFNGFQDTLYAHSNRQFYRD 259
Query: 383 CEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFIL-- 440
C+I GTIDFIFGN AAV QNCKI R PLP Q T+TAQG+K +Q+TG IQ S
Sbjct: 260 CDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQGKKDRNQNTGIIIQKSKFTPL 439
Query: 441 ---ATQPTYLGRPWKQYSRTVYINTYMSGMVQPRGWLEWFGDF-ALTTLWYGEYRNYGPG 496
T PTYLGRPWK +S TV + + + ++P GW+ W + ++T++Y EY+N GPG
Sbjct: 440 ENNLTAPTYLGRPWKDFSTTVIMQSDIGSFLKPVGWMSWVPNVEPVSTIFYAEYQNTGPG 619
Query: 497 ASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A ++ RVKW GY A FTVQ FI G WLP V+F + L
Sbjct: 620 ADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGPEWLPNAAVQFDSTL 760
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 244 bits (623), Expect = 7e-65
Identities = 115/198 (58%), Positives = 141/198 (71%), Gaps = 9/198 (4%)
Frame = +1
Query: 355 DQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQ 414
D S FY+CS EG+QDTLY HS RQFYREC IYGT+DFIFGN A VLQNC I+ R P P +
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNP-PNK 177
Query: 415 KVTVTAQGRKSPHQSTGFTIQDSFILATQP---------TYLGRPWKQYSRTVYINTYMS 465
T+TAQGR P+Q+TG +I +S + A TYLGRPWKQYSRTV++ TY+
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 466 GMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRF 525
G++ P GW+EW G+FAL TL+YGEY N GPG+S A RVKW GY ++ A+ AS F+V F
Sbjct: 358 GLINPAGWMEWSGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSVANF 537
Query: 526 IHGDSWLPGTGVKFTAGL 543
I G++WLP T V FT L
Sbjct: 538 IAGNAWLPSTKVPFTPSL 591
>CK606276
Length = 751
Score = 243 bits (621), Expect = 1e-64
Identities = 111/205 (54%), Positives = 145/205 (70%), Gaps = 12/205 (5%)
Frame = +1
Query: 351 RVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAP 410
R +D S FY CS EG+QDTLY HSLRQFY+ C+IYGT+DFIFGN AA+LQ+C +Y R P
Sbjct: 1 RNGADMSTFYNCSFEGYQDTLYVHSLRQFYKSCDIYGTVDFIFGNAAALLQDCNMYPRLP 180
Query: 411 LPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQ------------PTYLGRPWKQYSRTV 458
+ Q +TAQGR P+Q+TG +IQ+ I+A TYLGRPWK+YSRTV
Sbjct: 181 MQNQFNAITAQGRTDPNQNTGISIQNCCIIAASDLGDATNNYNGIKTYLGRPWKEYSRTV 360
Query: 459 YINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATAS 518
Y+ +++ G++ P+GW EW GDFAL+TL+Y E+ N+GPG++ + RV W GYH++ D A
Sbjct: 361 YMQSFIDGLIDPKGWNEWSGDFALSTLYYAEFANWGPGSNTSNRVAWEGYHLI-DEKDAD 537
Query: 519 YFTVQRFIHGDSWLPGTGVKFTAGL 543
FTV +FI G+ WLP TGV F AGL
Sbjct: 538 DFTVHKFIQGEKWLPQTGVPFXAGL 612
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 238 bits (606), Expect = 7e-63
Identities = 121/227 (53%), Positives = 157/227 (68%), Gaps = 10/227 (4%)
Frame = +2
Query: 248 GSGHYRTITEAVNAAPSHSNR-RYVIYVKKGLYRENVDMKRKMTNIMLVGDGIGQTIITS 306
GSG+++T+ +A+NAA + R+VI+VKKG+YREN+++ NIMLVGDG+ TIITS
Sbjct: 14 GSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTIITS 193
Query: 307 NRNFMQGWTTFRTATVAVSGKGFIARDMSFRNTAGPVNHQAVALRVDSDQSAFYRCSIEG 366
R+ G+TT+ +AT + G FIARD++F+N+AG QAVALR SD S FYRC I G
Sbjct: 194 ARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGIMG 373
Query: 367 FQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRKSP 426
+QDTL AH+ RQFYR+C IYGT+DFIFGN A V QNC I+ R PL Q +TAQGR P
Sbjct: 374 YQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRGDP 553
Query: 427 HQSTGFTIQDSFILATQP---------TYLGRPWKQYSRTVYINTYM 464
Q+TG +I +S I A T+LGRPW+QYSR V + T+M
Sbjct: 554 FQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMKTFM 694
>TC218157 similar to UP|Q8GUF8 (Q8GUF8) At2g47550/T30B22.15, partial (49%)
Length = 748
Score = 218 bits (554), Expect = 7e-57
Identities = 103/187 (55%), Positives = 125/187 (66%), Gaps = 8/187 (4%)
Frame = +1
Query: 365 EGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVTAQGRK 424
EG+QDTLY HSLRQFY EC GT+DFIFGN V QNC +Y R P+ Q +TAQGR
Sbjct: 1 EGYQDTLYVHSLRQFYSECXXXGTVDFIFGNAKVVFQNCNMYPRLPMSGQFNAITAQGRT 180
Query: 425 SPHQSTGFTIQDSFILATQP--------TYLGRPWKQYSRTVYINTYMSGMVQPRGWLEW 476
P+Q TG +I +S I A TYLGRPWK+YSRTVY+ T+M ++ +GW EW
Sbjct: 181 DPNQDTGISIHNSTIRAADDLASSNGVATYLGRPWKEYSRTVYMQTFMDSVIHAKGWREW 360
Query: 477 FGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDSWLPGTG 536
GDFAL+TL+Y EY N GPG+ RV WPGYH++ +A AS FTV F+ GD WLP TG
Sbjct: 361 DGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYHVI-NATDASNFTVSNFLLGDDWLPQTG 537
Query: 537 VKFTAGL 543
V +T L
Sbjct: 538 VSYTNNL 558
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 216 bits (551), Expect = 2e-56
Identities = 111/207 (53%), Positives = 134/207 (64%), Gaps = 9/207 (4%)
Frame = +3
Query: 346 QAVALRVDSDQSAFYRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKI 405
QAVALRV D SAF+ C I FQDTLY H+ RQF+ +C I GT+DFIFGN A V Q+C I
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 406 YTRAPLPLQKVTVTAQGRKSPHQSTGFTIQDSFILATQP---------TYLGRPWKQYSR 456
+ R P QK VTAQGR P+Q+TG IQ I AT TYLGRPWK+YSR
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 457 TVYINTYMSGMVQPRGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAAT 516
TV + + +S ++ P GW EW G+FAL+TL Y EY+N GPGA + RV W GY ++ DAA
Sbjct: 366 TVIMQSSISDVIDPIGWHEWSGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDAAE 545
Query: 517 ASYFTVQRFIHGDSWLPGTGVKFTAGL 543
A +T FI G SWL TG F+ GL
Sbjct: 546 ARDYTPGSFIGGSSWLGSTGFPFSLGL 626
>TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (69%)
Length = 747
Score = 215 bits (547), Expect = 5e-56
Identities = 95/189 (50%), Positives = 126/189 (66%), Gaps = 9/189 (4%)
Frame = +2
Query: 360 YRCSIEGFQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRAPLPLQKVTVT 419
Y+C +QD HS RQFYR+C++YGT+DF GN A VLQNC +Y R P Q+ T
Sbjct: 2 YKCXFVXYQDXXXVHSXRQFYRDCDVYGTVDFXXGNAATVLQNCNLYARKPNENQRNLFT 181
Query: 420 AQGRKSPHQSTGFTIQDSFILATQP---------TYLGRPWKQYSRTVYINTYMSGMVQP 470
AQGR+ P+Q+TG +I + + A YLGRPWK+YSRTVY+N+YM ++ P
Sbjct: 182 AQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGRPWKKYSRTVYLNSYMEDLIDP 361
Query: 471 RGWLEWFGDFALTTLWYGEYRNYGPGASLAGRVKWPGYHIMKDAATASYFTVQRFIHGDS 530
+GWLEW G FAL TL+YGEY N GPG++ + RV WPGY ++K+A A+ FTV+ FI G+
Sbjct: 362 KGWLEWNGTFALDTLYYGEYNNRGPGSNTSARVTWPGYRVIKNATEANQFTVRNFIQGNE 541
Query: 531 WLPGTGVKF 539
WL T + F
Sbjct: 542 WLSSTDIPF 568
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,929,123
Number of Sequences: 63676
Number of extensions: 303109
Number of successful extensions: 1545
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 1453
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1458
length of query: 545
length of database: 12,639,632
effective HSP length: 102
effective length of query: 443
effective length of database: 6,144,680
effective search space: 2722093240
effective search space used: 2722093240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0109.6


