

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.15
(312 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
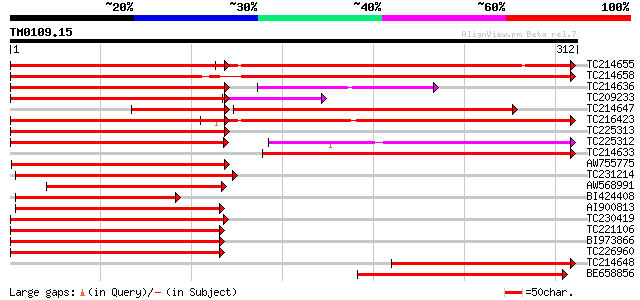
Score E
Sequences producing significant alignments: (bits) Value
TC214655 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 223 e-109
TC214658 homologue to UP|Q39856 (Q39856) Epoxide hydrolase, part... 384 e-107
TC214636 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 227 2e-69
TC209233 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 221 3e-66
TC214647 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 155 3e-62
TC216423 UP|O49857 (O49857) Epoxide hydrolase, complete 225 2e-59
TC225313 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partia... 213 9e-56
TC225312 similar to UP|Q76E11 (Q76E11) Soluble epoxide hydrolase... 181 2e-55
TC214633 similar to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragm... 193 1e-49
AW755775 similar to PIR|T07145|T07 epoxide hydrolase homolog - s... 189 1e-48
TC231214 similar to UP|Q9SD45 (Q9SD45) Epoxide hydrolase-like pr... 174 5e-44
AW568991 similar to PIR|T07145|T071 epoxide hydrolase homolog - ... 153 8e-38
BI424408 similar to PIR|T45731|T45 epoxide hydrolase-like protei... 133 1e-31
AI900813 weakly similar to PIR|T45731|T457 epoxide hydrolase-lik... 126 1e-29
TC230419 weakly similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase,... 119 1e-27
TC221106 similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partia... 119 2e-27
BI973866 weakly similar to GP|1354849|gb| epoxide hydrolase {Nic... 111 4e-25
TC226960 similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partia... 110 8e-25
TC214648 similar to UP|O49857 (O49857) Epoxide hydrolase, partia... 108 4e-24
BE658856 weakly similar to PIR|T07145|T07 epoxide hydrolase homo... 99 3e-21
>TC214655 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (90%)
Length = 1201
Score = 223 bits (569), Expect(2) = e-109
Identities = 101/121 (83%), Positives = 112/121 (92%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
MEGIEHRTVEVNGIKMHIAEKG+GPVVLFLHGFPELW+ W +QI+ L S GYHAVAPDLR
Sbjct: 7 MEGIEHRTVEVNGIKMHIAEKGEGPVVLFLHGFPELWHCWHNQIVALGSLGYHAVAPDLR 186
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDTD+P SI +YTCFHIV D+VALID LGA+QVFLVAHDWG+IIGWYLCMFRP++VKA
Sbjct: 187 GYGDTDAPPSIDSYTCFHIVADLVALIDSLGAEQVFLVAHDWGAIIGWYLCMFRPDKVKA 366
Query: 121 Y 121
Y
Sbjct: 367 Y 369
Score = 191 bits (484), Expect(2) = e-109
Identities = 115/199 (57%), Positives = 138/199 (68%), Gaps = 1/199 (0%)
Frame = +2
Query: 114 RPERVKAYAPSSE*NPTPE**IACVICMETITMSADFRNRAKWRLKWLKLALSMY*KTSL 173
RP + S+E PT E +AC +CME T+SADFRN+ KWRL+WLKLA SM+ KTSL
Sbjct: 362 RPMSASVFLSSAE-TPTSEPSMACALCMEKTTISADFRNQGKWRLRWLKLAQSMFSKTSL 538
Query: 174 QLEKLVLQSFPRESLELDSILIHLL-PCPLGSHKRILLIMSPSLRKRASLEG*TIIET*T 232
Q L LQSFP+ESLE D I I L+ P PLGS IL IM P++R RASL+ *TI E *T
Sbjct: 539 QFANLDLQSFPKESLEQD*IRICLMIPYPLGSQTMILPIMFPNMRNRASLDP*TITEI*T 718
Query: 233 *IGS*QHHGVECKSRCQ*SSSQVS*TWYTHHWVLRSISIVGVSRKMCQIWRKLLCKKGSV 292
*IGS*+HHG+ECKS+ +*++ QVS TWYT RS+S V VS KMC I+ K LC+K +
Sbjct: 719 *IGS*RHHGLECKSKWR*NT*QVSWTWYTPR*A*RSMSTVEVSNKMCPIY-KWLCRKE*L 895
Query: 293 TSIIKKQQMKLVATYTTSS 311
TSII K K TYT SS
Sbjct: 896 TSII*KLPRKSTITYTISS 952
>TC214658 homologue to UP|Q39856 (Q39856) Epoxide hydrolase, partial (87%)
Length = 1085
Score = 384 bits (985), Expect = e-107
Identities = 209/311 (67%), Positives = 236/311 (75%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
MEGIEHRTVEVNGIKMH+AEKG+GPVVLFLHGFPELWYSWRHQIL+LSS GY AVAPDLR
Sbjct: 16 MEGIEHRTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLR 195
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P S+++Y CFHIVGD+VALID LG +QVFLVAHDWG+II + +F E
Sbjct: 196 GYGDTEAPPSVSSYNCFHIVGDLVALIDSLGVQQVFLVAHDWGAII---VFLFSAE---- 354
Query: 121 YAPSSE*NPTPE**IACVICMETITMSADFRNRAKWRLKWLKLALSMY*KTSLQLEKLVL 180
T E *+ACV+CMET TMSADFRN+AKW+L+WLKLAL M+ KTSLQ+ KLVL
Sbjct: 355 -------TQTSER*MACVLCMETTTMSADFRNQAKWKLRWLKLALGMFSKTSLQVAKLVL 513
Query: 181 QSFPRESLELDSILIHLLPCPLGSHKRILLIMSPSLRKRASLEG*TIIET*T*IGS*QHH 240
SF ESLELDSI I L+P PLGS K ILLIMSP+ RK ASLE *T E T*IGS*Q H
Sbjct: 514 HSFRMESLELDSIQICLIPYPLGSQKMILLIMSPNSRKLASLEA*TTTEIST*IGS*QLH 693
Query: 241 GVECKSRCQ*SSSQVS*TWYTHHWVLRSISIVGVSRKMCQIWRKLLCKKGSVTSIIKKQQ 300
G ECKSRC *+S QVS T Y RSIS V S KMCQ K LC+K +TSIIKK
Sbjct: 694 GQECKSRCP*NSLQVSWTRYIPR*T*RSISTVEGSSKMCQT*NK*LCRKEWLTSIIKKPP 873
Query: 301 MKLVATYTTSS 311
K + T+T S
Sbjct: 874 RKSILTFTILS 906
>TC214636 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (58%)
Length = 778
Score = 227 bits (578), Expect(2) = 2e-69
Identities = 102/121 (84%), Positives = 114/121 (93%)
Frame = +3
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
MEG+ HRTVEVNGIKMHIAEKG+GPVVLFLHGFPELWYSWRHQIL+LSS GY AVAPDLR
Sbjct: 60 MEGVIHRTVEVNGIKMHIAEKGEGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLR 239
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P SI +YTCFHIVGD+VALID LG +QVFLVAHDWG+++GWYLCMFRPE+VKA
Sbjct: 240 GYGDTEAPPSIDSYTCFHIVGDLVALIDSLGVQQVFLVAHDWGALMGWYLCMFRPEKVKA 419
Query: 121 Y 121
Y
Sbjct: 420 Y 422
Score = 53.5 bits (127), Expect(2) = 2e-69
Identities = 43/100 (43%), Positives = 58/100 (58%)
Frame = +1
Query: 137 CVICMETITMSADFRNRAKWRLKWLKLALSMY*KTSLQLEKLVLQSFPRESLELDSILIH 196
C ME T SAD R++A+ ++ K+AL+ *KT + L Q + R+ E SI I
Sbjct: 481 CGPYMEMTTTSADSRSQARQKVS*PKIALNR**KTYS*VANLGHQFWRRK--EWVSIPIL 654
Query: 197 LLPCPLGSHKRILLIMSPSLRKRASLEG*TIIET*T*IGS 236
P PLGSHK+ M +L+++ASLE T IE T*IGS
Sbjct: 655 QCPFPLGSHKKTSRTMLLNLKRQASLEDSTTIEIST*IGS 774
>TC209233 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (53%)
Length = 590
Score = 221 bits (563), Expect(2) = 3e-66
Identities = 97/121 (80%), Positives = 111/121 (91%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEHRTVEVNGIKMH+AEKG+G VVLFLHGFPELWYSWRHQIL LSS GY AVAPDLR
Sbjct: 43 MESIEHRTVEVNGIKMHVAEKGEGAVVLFLHGFPELWYSWRHQILALSSLGYRAVAPDLR 222
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P ++ +YTCFH+VGDI+ALID LG +VFLVAHDWG+IIGWYLC+FRP+R+KA
Sbjct: 223 GYGDTEAPSTVNSYTCFHLVGDIIALIDSLGVDKVFLVAHDWGAIIGWYLCLFRPDRIKA 402
Query: 121 Y 121
Y
Sbjct: 403 Y 405
Score = 48.5 bits (114), Expect(2) = 3e-66
Identities = 26/57 (45%), Positives = 34/57 (59%)
Frame = +2
Query: 118 VKAYAPSSE*NPTPE**IACVICMETITMSADFRNRAKWRLKWLKLALSMY*KTSLQ 174
V APSSE P + + CME T SAD RN+ +W+L+WL L L + *+T LQ
Sbjct: 416 VSPSAPSSEETPNRKPLTFSIPCMEMTTTSADSRNQGRWKLRWLALTLRI**RTYLQ 586
>TC214647 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (60%)
Length = 659
Score = 155 bits (393), Expect(2) = 3e-62
Identities = 91/156 (58%), Positives = 108/156 (68%)
Frame = +1
Query: 124 SSE*NPTPE**IACVICMETITMSADFRNRAKWRLKWLKLALSMY*KTSLQLEKLVLQSF 183
SS PT E *+ACV+CME S DFRN+ KWRL+WLKLALSM+ KTSLQL KLVL+ F
Sbjct: 184 SSPETPTSEP*MACVLCMEKTITSPDFRNQGKWRLRWLKLALSMFSKTSLQLAKLVLRPF 363
Query: 184 PRESLELDSILIHLLPCPLGSHKRILLIMSPSLRKRASLEG*TIIET*T*IGS*QHHGVE 243
PRES ELDSI I + C LGS +I MS +L K ASLE *T + T IGS*+HHG+E
Sbjct: 364 PRESTELDSIRI*PISCLLGSQMKI*SYMSRNLIKPASLEA*TTTQISTQIGS*RHHGLE 543
Query: 244 CKSRCQ*SSSQVS*TWYTHHWVLRSISIVGVSRKMC 279
+S C *+S QV+ TW T + RSIS VS+K C
Sbjct: 544 LQSMCL*NSLQVNWTWNTLAYESRSISTAVVSQKTC 651
Score = 100 bits (250), Expect(2) = 3e-62
Identities = 43/54 (79%), Positives = 50/54 (91%)
Frame = +3
Query: 68 PISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAY 121
P SI +YTCFHIVGD+VALID LG +QVFLVAHDWG+++GWYLCMFRPE+VKAY
Sbjct: 3 PPSIDSYTCFHIVGDLVALIDSLGVQQVFLVAHDWGALMGWYLCMFRPEKVKAY 164
>TC216423 UP|O49857 (O49857) Epoxide hydrolase, complete
Length = 1343
Score = 225 bits (573), Expect = 2e-59
Identities = 101/121 (83%), Positives = 114/121 (93%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME I+HRTVEVNGIKMH+AEKG+GPVVLFLHGFPELWYSWRHQIL+LSS GY AVAPDLR
Sbjct: 148 MEQIKHRTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLR 327
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P SI++Y CFHIVGD+VALID LG +QVFLVAHDWG+IIGWYLCMFRP++VKA
Sbjct: 328 GYGDTEAPPSISSYNCFHIVGDLVALIDSLGVQQVFLVAHDWGAIIGWYLCMFRPDKVKA 507
Query: 121 Y 121
Y
Sbjct: 508 Y 510
Score = 196 bits (498), Expect = 1e-50
Identities = 121/210 (57%), Positives = 138/210 (65%), Gaps = 4/210 (1%)
Frame = +2
Query: 106 IGWYLCM----FRPERVKAYAPSSE*NPTPE**IACVICMETITMSADFRNRAKWRLKWL 161
+G Y C RP S+E T E +ACV+CMET TMSADFRN+ KWRL+WL
Sbjct: 467 VGIYACFALTKLRPMSASVSLSSAE-TQTSERWMACVLCMETTTMSADFRNQGKWRLRWL 643
Query: 162 KLALSMY*KTSLQLEKLVLQSFPRESLELDSILIHLLPCPLGSHKRILLIMSPSLRKRAS 221
KLALSM+ KTSLQL LVLQ FPRE +SI PCPLGS K+I IMSP+LRK S
Sbjct: 644 KLALSMFSKTSLQLAILVLQFFPREGF--NSIQKCPTPCPLGSQKKISPIMSPNLRKPDS 817
Query: 222 LEG*TIIET*T*IGS*QHHGVECKSRCQ*SSSQVS*TWYTHHWVLRSISIVGVSRKMCQI 281
L+ *T E T*IGS*+HHG E KSRC *++ QVS TWYT RSIS S KMCQI
Sbjct: 818 LDP*TTTEIST*IGS*RHHGQEGKSRCP*NT*QVSWTWYTTR*T*RSISTAEGSSKMCQI 997
Query: 282 WRKLLCKKGSVTSIIKKQQMKLVATYTTSS 311
K LC+K +TSIIKKQQ K + TYT S
Sbjct: 998 *NK*LCRKEWLTSIIKKQQRKSIITYTILS 1087
>TC225313 similar to UP|Q39856 (Q39856) Epoxide hydrolase, partial (51%)
Length = 1307
Score = 213 bits (542), Expect = 9e-56
Identities = 95/121 (78%), Positives = 108/121 (88%)
Frame = +3
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
MEGI HRTVEVNGIKMH+AEKG+GPVVLFLHGFPELWYSWRHQIL LS+ GY AVAPDLR
Sbjct: 627 MEGIVHRTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQILALSNLGYRAVAPDLR 806
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P SI++YT H+V DIVALI L QVFLVAHDWG++IGWYLC+FRP+R+KA
Sbjct: 807 GYGDTEAPASISSYTILHLVSDIVALIHSLAVDQVFLVAHDWGAVIGWYLCLFRPDRIKA 986
Query: 121 Y 121
Y
Sbjct: 987 Y 989
>TC225312 similar to UP|Q76E11 (Q76E11) Soluble epoxide hydrolase, partial
(96%)
Length = 1213
Score = 181 bits (460), Expect(2) = 2e-55
Identities = 78/120 (65%), Positives = 97/120 (80%)
Frame = +2
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH+TV NGI MH+A G GPVVLFLHGFPELWY+WRHQ+L+LS+ GY A+APDLR
Sbjct: 41 MEKIEHKTVRTNGINMHVASIGSGPVVLFLHGFPELWYTWRHQLLSLSAVGYRAIAPDLR 220
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDTD+P ++Y+ HIV D+V L+D LG ++VFLV HDWG+ I W+ C+ RP+RVKA
Sbjct: 221 GYGDTDAPPDASSYSALHIVADLVGLLDALGIERVFLVGHDWGASIAWHFCLLRPDRVKA 400
Score = 52.0 bits (123), Expect(2) = 2e-55
Identities = 58/172 (33%), Positives = 82/172 (46%), Gaps = 3/172 (1%)
Frame = +3
Query: 143 TITMSADFRNRAKWRLKWLKLALSMY*KTSLQL---EKLVLQSFPRESLELDSILIHLLP 199
TIT+SA RN + + L L *+ L L + LV + L +LI+L
Sbjct: 480 TITISAGSRNLGRLKKSLHVLVLREL*RHLLHLAIHDHLVCLRR*DLVVHLTFVLIYLRG 659
Query: 200 CPLGSHKRILLIMSPSLRKRASLEG*TIIET*T*IGS*QHHGVECKSRCQ*SSSQVS*TW 259
P K++ +IM +L ++ SL *T I T* GS Q HG+E + R Q S + T
Sbjct: 660 *P----KKMSIIMPANLNRKVSLAA*TTIALWT*PGSSQQHGLEYRLRYQSSLLWETLTL 827
Query: 260 YTHHWVLRSISIVGVSRKMCQIWRKLLCKKGSVTSIIKKQQMKLVATYTTSS 311
+T VLRSI I+ R+ + L K +T I+K + YTTSS
Sbjct: 828 HTTLQVLRSIYIMVALRETYHSCKNWL*WKE*LTL*IRKGPKRSARIYTTSS 983
>TC214633 similar to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragment) ,
partial (63%)
Length = 722
Score = 193 bits (490), Expect = 1e-49
Identities = 111/172 (64%), Positives = 124/172 (71%)
Frame = +1
Query: 140 CMETITMSADFRNRAKWRLKWLKLALSMY*KTSLQLEKLVLQSFPRESLELDSILIHLLP 199
CME S DFRN+ KWRL+WLKLALSM+ KTSLQL KLVL+ FPRES ELDSI I +
Sbjct: 1 CMEKTITSPDFRNQGKWRLRWLKLALSMFSKTSLQLAKLVLRPFPRESTELDSIRI*PIS 180
Query: 200 CPLGSHKRILLIMSPSLRKRASLEG*TIIET*T*IGS*QHHGVECKSRCQ*SSSQVS*TW 259
C LGS K+I IMSP+LRK ASLE *T E T IGS*+HHG ECKSRC *+S QV+ TW
Sbjct: 181 CLLGSQKKI*PIMSPNLRKPASLEA*TTTEISTQIGS*RHHGQECKSRCL*NSLQVNWTW 360
Query: 260 YTHHWVLRSISIVGVSRKMCQIWRKLLCKKGSVTSIIKKQQMKLVATYTTSS 311
YT RSIS V VSRK C+I K LC+K +TSIIKKQQ V TYT S
Sbjct: 361 YTPR*ESRSISTVEVSRKTCRI*NK*LCRKELLTSIIKKQQRISVITYTILS 516
>AW755775 similar to PIR|T07145|T07 epoxide hydrolase homolog - soybean,
partial (41%)
Length = 469
Score = 189 bits (480), Expect = 1e-48
Identities = 83/120 (69%), Positives = 101/120 (84%)
Frame = +2
Query: 2 EGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
EGI H+TVEVNGIKMH+AEKG+GPVVLFLHGFPELWYSWRHQIL ++ G+ AVA +LRG
Sbjct: 2 EGIVHKTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQILAFNNLGFRAVASELRG 181
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAY 121
+ DT++P SI++YT H++ DI ALI L QVFLV HDWG+ IGWYLC+FRP+R+KAY
Sbjct: 182 FRDTEAPASISSYTILHLISDIGALIHSLAVDQVFLVTHDWGAGIGWYLCLFRPDRIKAY 361
>TC231214 similar to UP|Q9SD45 (Q9SD45) Epoxide hydrolase-like protein
(AT3g51000/F24M12_40), partial (73%)
Length = 1104
Score = 174 bits (441), Expect = 5e-44
Identities = 72/122 (59%), Positives = 94/122 (77%)
Frame = +2
Query: 4 IEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYG 63
++H+ ++ NGI +H+AEKG GP+VL LHGFPE WY+WRHQI L+ GYH VAPDLRGYG
Sbjct: 77 VKHQRIKTNGIWLHVAEKGTGPLVLLLHGFPETWYAWRHQINFLAHHGYHVVAPDLRGYG 256
Query: 64 DTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAYAP 123
D+DSPI ++YT H+VGDI+ +DH G Q F+V DWG++IGW+L +FRPERVK +
Sbjct: 257 DSDSPIDPSSYTIHHLVGDIIGFLDHFGQHQAFIVGSDWGAVIGWHLSLFRPERVKGFVA 436
Query: 124 SS 125
S
Sbjct: 437 LS 442
>AW568991 similar to PIR|T07145|T071 epoxide hydrolase homolog - soybean,
partial (29%)
Length = 301
Score = 153 bits (387), Expect = 8e-38
Identities = 66/99 (66%), Positives = 80/99 (80%)
Frame = +3
Query: 21 KGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYGDTDSPISITTYTCFHIV 80
KG+GPVVLFLH FPELWYSW HQIL+L S YH + P+L GY DT +P SI++Y CFHIV
Sbjct: 3 KGEGPVVLFLHSFPELWYSWHHQILSLXSLSYHTITPNLHGYSDTKTPPSISSYNCFHIV 182
Query: 81 GDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
GD++ALID LG +Q+FLVAHDWG+II YL MF P ++K
Sbjct: 183 GDLIALIDSLGVQQMFLVAHDWGAIIS*YLYMFHPNKIK 299
>BI424408 similar to PIR|T45731|T45 epoxide hydrolase-like protein -
Arabidopsis thaliana, partial (33%)
Length = 421
Score = 133 bits (334), Expect = 1e-31
Identities = 55/91 (60%), Positives = 71/91 (77%)
Frame = +2
Query: 4 IEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYG 63
++H+ ++ NGI +H+AEKG GP+VL LHGFPE WY+WRHQI L+ GYH VAPDLRGYG
Sbjct: 77 VKHQRIKTNGIWLHVAEKGTGPLVLLLHGFPETWYAWRHQINFLAHHGYHVVAPDLRGYG 256
Query: 64 DTDSPISITTYTCFHIVGDIVALIDHLGAKQ 94
D+DSPI ++YT H+VGDI+ +DH G Q
Sbjct: 257 DSDSPIDPSSYTIHHLVGDIIGFLDHFGQHQ 349
>AI900813 weakly similar to PIR|T45731|T457 epoxide hydrolase-like protein -
Arabidopsis thaliana, partial (28%)
Length = 406
Score = 126 bits (316), Expect = 1e-29
Identities = 56/115 (48%), Positives = 79/115 (68%)
Frame = +2
Query: 4 IEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRGYG 63
++H+ ++ N I +H+A+KG +VL LHGFP+ +WRHQI L+ GYH VAPDL GYG
Sbjct: 2 VKHQRIKTNWIWLHVAKKGTLSLVLMLHGFPQTCDAWRHQINFLAHHGYHVVAPDLIGYG 181
Query: 64 DTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERV 118
D+DS I ++YT H+V DI+ +DH G Q F+ D G++IGW L +F P+RV
Sbjct: 182 DSDSAIDRSSYTIHHLVCDIIGFLDHSGRHQAFIAGSDCGAVIGWPLNLFMPDRV 346
>TC230419 weakly similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partial
(35%)
Length = 536
Score = 119 bits (299), Expect = 1e-27
Identities = 57/121 (47%), Positives = 78/121 (64%), Gaps = 1/121 (0%)
Frame = +2
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGP-VVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
M I+H VEV G+K+H+AE G G V+FLHGFPE+WY+WRHQ++ ++ GY A+A D
Sbjct: 110 MAKIQHSEVEVKGLKLHVAEIGSGSKAVVFLHGFPEIWYTWRHQMIAAANAGYRAIAFDF 289
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
RGYG ++ P T + +V +IV L+D L Q FLV D+G+I G+ PERV
Sbjct: 290 RGYGLSEHPAEPEKETMYDLVDEIVGLLDALNITQAFLVGKDFGAIPGYLTAAVHPERVA 469
Query: 120 A 120
A
Sbjct: 470 A 472
>TC221106 similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partial (31%)
Length = 585
Score = 119 bits (298), Expect = 2e-27
Identities = 57/119 (47%), Positives = 78/119 (64%), Gaps = 1/119 (0%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGP-VVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
ME I+H VEV G+K+H+AE G G V+FLHGFPE+WY+WRHQ++++++ GY A+A D
Sbjct: 13 MEKIQHSEVEVKGLKLHVAEIGSGSKTVVFLHGFPEIWYTWRHQMISVANAGYRAIAFDF 192
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERV 118
RGYG + P T F +V +IV L+D L + FLV D+G+I G PERV
Sbjct: 193 RGYGLSQQPAEPEKETMFDLVHEIVGLLDALNISKAFLVGKDFGAIPGHLTTAVHPERV 369
>BI973866 weakly similar to GP|1354849|gb| epoxide hydrolase {Nicotiana
tabacum}, partial (33%)
Length = 420
Score = 111 bits (278), Expect = 4e-25
Identities = 53/119 (44%), Positives = 77/119 (64%), Gaps = 1/119 (0%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGP-VVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
M+ IEH+ V V +K+H+AE G G V+FLHGFPE+WYSWRHQ++ L+ G+ AV+ D
Sbjct: 13 MDRIEHKFVNVGDLKLHVAEIGSGGNAVVFLHGFPEIWYSWRHQMIALADAGFRAVSFDY 192
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERV 118
RGYG +D P T F ++ D++ ++D L +VFLV D+G+ + + PERV
Sbjct: 193 RGYGLSDPPPPGNKATWFDLLNDLLHILDALALSKVFLVGKDFGARPAHFFSILHPERV 369
>TC226960 similar to UP|Q9ZP87 (Q9ZP87) Epoxide hydrolase, partial (72%)
Length = 1201
Score = 110 bits (275), Expect = 8e-25
Identities = 51/119 (42%), Positives = 77/119 (63%), Gaps = 1/119 (0%)
Frame = +1
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGP-VVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
M+ I+H+ V V +K+H+AE G GP V+FLHGFPE+WYSWRHQ++ L+ G+ AV+ D
Sbjct: 55 MDPIQHKFVNVGALKLHVAETGTGPNAVVFLHGFPEIWYSWRHQMIALAGAGFRAVSFDY 234
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERV 118
RGYG +D P + ++ D++ ++D L +VFLV D+G+ + + PERV
Sbjct: 235 RGYGLSDPPPEPDKTSWSDLLSDLLHILDALALSKVFLVGKDFGARPAYLFSILHPERV 411
>TC214648 similar to UP|O49857 (O49857) Epoxide hydrolase, partial (30%)
Length = 682
Score = 108 bits (269), Expect = 4e-24
Identities = 64/101 (63%), Positives = 71/101 (69%)
Frame = +2
Query: 211 IMSPSLRKRASLEG*TIIET*T*IGS*QHHGVECKSRCQ*SSSQVS*TWYTHHWVLRSIS 270
IMSP+LRK ASLE *T E T IGS*+HHG ECKSRC *+S QV+ TWYT RSIS
Sbjct: 152 IMSPNLRKPASLEA*TTTEISTQIGS*RHHGQECKSRCL*NSLQVNWTWYTPR*ESRSIS 331
Query: 271 IVGVSRKMCQIWRKLLCKKGSVTSIIKKQQMKLVATYTTSS 311
V VSRK C+I K LC+K +TSIIKKQQ V TYT S
Sbjct: 332 TVEVSRKTCRI*NK*LCRKELLTSIIKKQQRISVITYTILS 454
>BE658856 weakly similar to PIR|T07145|T07 epoxide hydrolase homolog -
soybean, partial (43%)
Length = 567
Score = 98.6 bits (244), Expect = 3e-21
Identities = 62/116 (53%), Positives = 73/116 (62%)
Frame = -3
Query: 192 SILIHLLPCPLGSHKRILLIMSPSLRKRASLEG*TIIET*T*IGS*QHHGVECKSRCQ*S 251
SI I P PLGSHK+ M +L+++ASLE T IE T*IGS QHHG+E KS+ Q*S
Sbjct: 481 SIPILQCPFPLGSHKKTSRTMLLNLKRQASLEDSTTIEIST*IGSSQHHGLEHKSKYQ*S 302
Query: 252 SSQVS*TWYTHHWVLRSISIVGVSRKMCQIWRKLLCKKGSVTSIIKKQQMKLVATY 307
S QV YT HW R+I +SRKM QIWRK LCKKG +T KK Q T+
Sbjct: 301 SLQVIWI*YTLHWGRRTILRAVLSRKMYQIWRK*LCKKGWLTLAPKKLQRMSAITF 134
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.340 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,147,754
Number of Sequences: 63676
Number of extensions: 273251
Number of successful extensions: 2121
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 2096
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2105
length of query: 312
length of database: 12,639,632
effective HSP length: 97
effective length of query: 215
effective length of database: 6,463,060
effective search space: 1389557900
effective search space used: 1389557900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0109.15


