

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
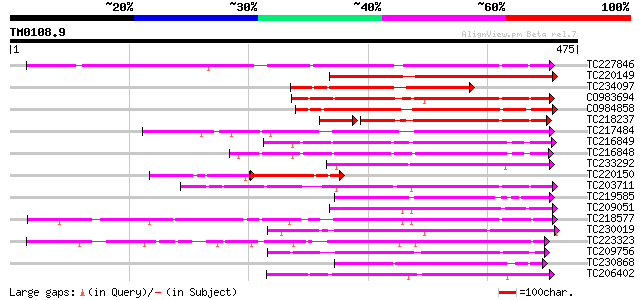
Query= TM0108.9
(475 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 263 2e-70
TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferas... 249 2e-66
TC234097 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin 5... 210 1e-54
CO983694 189 3e-48
CO984858 183 1e-46
TC218237 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferas... 142 6e-39
TC217484 153 2e-37
TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransf... 151 5e-37
TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 8... 142 3e-34
TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, pa... 140 9e-34
TC220150 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin 5... 93 2e-33
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 134 7e-32
TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltr... 134 9e-32
TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 134 1e-31
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 131 6e-31
TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferas... 129 3e-30
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 127 1e-29
TC209756 126 2e-29
TC230868 weakly similar to UP|Q94AB5 (Q94AB5) AT3g46660/F12A12_1... 125 4e-29
TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, p... 124 9e-29
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 263 bits (671), Expect = 2e-70
Identities = 157/449 (34%), Positives = 238/449 (52%), Gaps = 7/449 (1%)
Frame = +2
Query: 15 LIVSYPGQGQINPTLQFSKRLIAMGAHVTIVTTLHMLRRMTTKTTIPGLSYTAFSDGFDD 74
L++ YP QG INP +QF+KRL + G T+ TT + + P ++ A SDGFD
Sbjct: 2 LVLPYPAQGHINPLVQFAKRLASKGVKATVATTHYTANSINA----PNITVEAISDGFDQ 169
Query: 75 GFIVDVATHFNLYTPELKRRGSEFITNLILFAENEGHPFTCLIHTLFVPWAPQVARELNL 134
+ L+ + GS ++ LI + P TC+++ F PW VA++ +
Sbjct: 170 AGFAQTNNNVQLFLASFRTNGSRTLSELIRKHQQTPSPSTCIVYDSFFPWVLDVAKQHGI 349
Query: 135 ITAKLWIEPTTVMSILYNYFLGGDYIAKKNT-----VESSCSIEFSGLP-FSLSPHDMPS 188
A + V +I G + K V ++ LP F P P+
Sbjct: 350 YGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLRVPGLPPLDSRALPSFVRFPESYPA 529
Query: 189 YLISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALEPEAPRAVDKLNMIP-IGLLIPSA 247
Y+ K S L D+ + VNTFEALE E + + +L IG ++PS
Sbjct: 530 YMAMKLSQFSNLNNADW----------MFVNTFEALESEVLKGLTELFPAKMIGPMVPSG 679
Query: 248 FLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARAL 307
+LD + D +G + + + + WL+SKPP SVVY+SFGS + L++ QM+E+A L
Sbjct: 680 YLDGR-IKGDKGYGASLWKPLTEECSNWLESKPPQSVVYISFGSMVSLTEEQMEEVAWGL 856
Query: 308 LDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLT 367
+ FLWV+RE E + LG E +++ G I WC+Q+E+L+H++ GCF+T
Sbjct: 857 KESGVSFLWVLRESE-------HGKLPLGYRESVKDKGLIVTWCNQLELLAHQATGCFVT 1015
Query: 368 HCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRR 427
HCGWNST+ESL VP+V PQW DQ +AK ++++W++GV +E GIV +E +
Sbjct: 1016HCGWNSTLESLSLGVPVVCLPQWADQLPDAKFLDEIWEVGVWP--KEDEKGIVRKQEFVQ 1189
Query: 428 GLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
L VM+ G++ +E+RRNA KWK LA+EA
Sbjct: 1190SLKDVME-GQRSQEIRRNANKWKKLAREA 1273
>TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferase like
protein, partial (27%)
Length = 820
Score = 249 bits (636), Expect = 2e-66
Identities = 127/192 (66%), Positives = 148/192 (76%), Gaps = 1/192 (0%)
Frame = +2
Query: 269 SNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGE 328
SNDY+EWLDS+P SVVYVSFG+ VL+ QM E+ARALLD + FLWVIR+ + +D
Sbjct: 5 SNDYVEWLDSQPELSVVYVSFGTLAVLADRQMKELARALLDSGYLFLWVIRDMQGIEDNC 184
Query: 329 KEKEEELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFP 388
+E ELE+ GKI KWCSQVEVLSH S+GCF+THCGWNSTMESL S VPMVAFP
Sbjct: 185 RE---------ELEQRGKIVKWCSQVEVLSHGSLGCFVTHCGWNSTMESLGSGVPMVAFP 337
Query: 389 QWTDQNTNAKLIEDVWKIGVRVDLHVN-EDGIVEGEEIRRGLTVVMKNGEKGEELRRNAK 447
QWTDQ TNAK+++DVWK GVRVD VN E+GIVE EEIR+ L VVM +G KG+E RRNA
Sbjct: 338 QWTDQGTNAKMVQDVWKTGVRVDDKVNVEEGIVEAEEIRKCLDVVMGSGGKGQEFRRNAD 517
Query: 448 KWKGLAKEAGKE 459
KWK LA+EA E
Sbjct: 518 KWKCLAREAVTE 553
>TC234097 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin
5-O-glucosyltransferase, partial (30%)
Length = 435
Score = 210 bits (535), Expect = 1e-54
Identities = 112/154 (72%), Positives = 119/154 (76%)
Frame = +1
Query: 236 NMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVL 295
NMIPIG LIPSAFLD KD DTSFGGDI + PSND EWLDSKP SVVYVSFGS+ VL
Sbjct: 7 NMIPIGPLIPSAFLDGKD-PTDTSFGGDIFR-PSNDCGEWLDSKPEMSVVYVSFGSFCVL 180
Query: 296 SKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVE 355
SK QM+E+A ALLDC PFLWV REKE EEEL EELE+ GKI WCSQVE
Sbjct: 181 SKKQMEELALALLDCGSPFLWVSREKE---------EEELSCREELEQKGKIVNWCSQVE 333
Query: 356 VLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQ 389
VLSHRS+GCF+THCGWNSTMESL S VPM AFPQ
Sbjct: 334 VLSHRSVGCFVTHCGWNSTMESLASGVPMFAFPQ 435
>CO983694
Length = 858
Score = 189 bits (479), Expect = 3e-48
Identities = 108/222 (48%), Positives = 139/222 (61%), Gaps = 2/222 (0%)
Frame = -1
Query: 237 MIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLS 296
++ IG +P LD K +DT + Q S+ I WL KP SV+Y+SFGS + S
Sbjct: 858 ILMIGPTVPXXHLD-KAVPNDTDNXXNXFQVDSSA-ISWLRQKPAGSVIYISFGSMVCFS 685
Query: 297 KTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGK--IAKWCSQV 354
QM+EIA L+ FLWVI + E+ K +ELG EE+ G+ I W Q+
Sbjct: 684 SQQMEEIALGLMATGFNFLWVIPDLER-----KNLPKELG--EEINACGRGLIVNWTPQL 526
Query: 355 EVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHV 414
EVLS+ ++GCF THCGWNST+E+L VPMVA PQWTDQ TNAK +EDVWK+G+RV
Sbjct: 525 EVLSNHAVGCFFTHCGWNSTLEALCLGVPMVALPQWTDQPTNAKFVEDVWKVGIRV--KE 352
Query: 415 NEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
NE+GIV EE+ + VVM+ + G E+R NAKKWK LA EA
Sbjct: 351 NENGIVTREEVENCIRVVMEK-DLGREMRINAKKWKELAIEA 229
>CO984858
Length = 778
Score = 183 bits (465), Expect = 1e-46
Identities = 97/220 (44%), Positives = 137/220 (62%)
Frame = -2
Query: 240 IGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQ 299
IG +PS FLD K D +G + Q S + +EWLD KP SVVYVSFGS +S+ Q
Sbjct: 774 IGPNVPSFFLD-KQCEDDQDYG--VTQFKSEECVEWLDDKPKGSVVYVSFGSMATMSEEQ 604
Query: 300 MDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSH 359
M+E+A L +C FLWV+R E+ + + + E++ E G + WCSQ++VL+H
Sbjct: 603 MEEVACCLRECSSYFLWVVRASEEIKLPK--------DFEKITEKGLVVTWCSQLKVLAH 448
Query: 360 RSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGI 419
++GCF+THCGWNS +E+L VP +A P W+DQ TNAKLI DVWKIG+R V+E I
Sbjct: 447 EAVGCFVTHCGWNSILETLCLGVPTIAIPCWSDQRTNAKLIADVWKIGIRTP--VDEKNI 274
Query: 420 VEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
V E ++ + +M ++ +E++ NA +WK LA A E
Sbjct: 273 VRREALKHCIKEIM---DRDKEMKTNAIQWKTLAVRATAE 163
>TC218237 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (57%)
Length = 1176
Score = 142 bits (358), Expect(2) = 6e-39
Identities = 75/160 (46%), Positives = 104/160 (64%)
Frame = +3
Query: 295 LSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQV 354
L++ Q +E+A L D F+WVIR+ +K G+ KE + E G I WC Q+
Sbjct: 141 LNEEQTEELAWGLGDSGSYFMWVIRDCDK---GKLPKE-----FADTSEKGLIVSWCPQL 296
Query: 355 EVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHV 414
+VL+H ++GCFLTHCGWNST+E+L VP++A P WTDQ TNAKL++DVWKIGV+
Sbjct: 297 QVLTHEALGCFLTHCGWNSTLEALSLGVPVIAMPLWTDQITNAKLLKDVWKIGVKA--VA 470
Query: 415 NEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAK 454
+E IV E I + +++ EKG E+++NA KWK LAK
Sbjct: 471 DEKEIVRRETITHCIKEILET-EKGNEIKKNAIKWKNLAK 587
Score = 37.0 bits (84), Expect(2) = 6e-39
Identities = 17/32 (53%), Positives = 21/32 (65%)
Frame = +2
Query: 260 FGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGS 291
+G ++ S I+WLD KP SVVYVSFGS
Sbjct: 35 YGVNMYNPNSEACIKWLDEKPKGSVVYVSFGS 130
>TC217484
Length = 1651
Score = 153 bits (386), Expect = 2e-37
Identities = 111/354 (31%), Positives = 175/354 (49%), Gaps = 9/354 (2%)
Frame = +2
Query: 112 PFTCLIHTLFVPWAPQVARELNLITAKLWIEPTTVMSILYNYFLGGDY---IAKKNTVES 168
P T ++ + + W VA N+ A W + S+L++ + + K+T++
Sbjct: 197 PPTAILGCVELRWPIAVANRRNIPVAAFWTMSASFYSMLHHLDVFARHRGLTVDKDTMDG 376
Query: 169 SCSIEFSGLPFSLSPH--DMPSYLISKGSYVLPLFKEDFKELRG*ANTTIL--VNTFEAL 224
+P S H D+ + L V+ L E ++ AN +L V EA
Sbjct: 377 QAE----NIPGISSAHLADLRTVLHENDQRVMQLALECISKVPR-ANYLLLTTVQELEAE 541
Query: 225 EPEAPRAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSV 284
E+ +A+ + PIG IP L + +D S +DYI+WLDS+PP SV
Sbjct: 542 TIESLKAIFPFPVYPIGPAIPYLELGQNPLNNDHS----------HDYIKWLDSQPPESV 691
Query: 285 VYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEV 344
+Y+SFGS++ +S TQMD+I AL +LWV R F E+ +
Sbjct: 692 LYISFGSFLSVSTTQMDQIVEALNSSEVRYLWVARANASFLK------------EKCGDK 835
Query: 345 GKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVW 404
G + WC Q++VLSH S+G F +HCGWNST+E+L + VPM+ FP + DQ N+ I D W
Sbjct: 836 GMVVPWCDQLKVLSHSSVGGFWSHCGWNSTLEALFAGVPMLTFPLFLDQVPNSSQIVDEW 1015
Query: 405 KIGVRVDL-HVNEDGIVEGEEIRRGLTVVMK-NGEKGEELRRNAKKWKGLAKEA 456
K G +V+ ++ + IV E+I + M ++G+E+R A++ K + A
Sbjct: 1016KNGSKVETSKLDSEVIVAKEKIEELVKRFMDLQSQEGKEIRDRAREIKVMCLRA 1177
>TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8,
partial (47%)
Length = 1125
Score = 151 bits (382), Expect = 5e-37
Identities = 94/248 (37%), Positives = 139/248 (55%), Gaps = 2/248 (0%)
Frame = +2
Query: 213 NTTILVNTFEALEPEAPRAVDKL--NMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSN 270
NTTIL NTF+ LE + A+ + ++ PIG L++ +H TS G ++
Sbjct: 161 NTTILFNTFDGLESDVMNALSSMFPSLYPIGPF--PLLLNQSPQSHLTSLGSNLWNEDL- 331
Query: 271 DYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKE 330
+ +EWL+SK SVVYV+FGS V+S Q+ E A L + + PFLW+IR + G
Sbjct: 332 ECLEWLESKESRSVVYVNFGSITVMSAEQLLEFAWGLANSKKPFLWIIRP-DLVIGGSVI 508
Query: 331 KEEELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQW 390
E + E + IA WC Q +VL+H S+G FLTHCGWNST ES+ + VPM+ +P +
Sbjct: 509 LSSEF--VSETRDRSLIASWCPQEQVLNHPSIGVFLTHCGWNSTTESVCAGVPMLCWPFF 682
Query: 391 TDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWK 450
+Q TN + I + W+IG+ +D + E E++ L V GEKG+++R + K
Sbjct: 683 AEQPTNCRYICNEWEIGMEIDTSAKRE---EVEKLVNELMV----GEKGKKMREKVMELK 841
Query: 451 GLAKEAGK 458
A+E K
Sbjct: 842 RKAEEVTK 865
>TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8, partial
(39%)
Length = 1113
Score = 142 bits (358), Expect = 3e-34
Identities = 95/275 (34%), Positives = 150/275 (54%), Gaps = 4/275 (1%)
Frame = +1
Query: 185 DMPSYL--ISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALEPEAPRAVDKL--NMIPI 240
D+P +L ++L F E +++ + + + NTF LE +A A+ + ++ I
Sbjct: 40 DLPDFLRTTDPNDFMLHFFIEVAEKVP--SASAVAFNTFHELERDAINALPSMFPSLYSI 213
Query: 241 GLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQM 300
G +FLD+ S G ++ + + ++WL+SK P SVVYV+FGS V+S Q+
Sbjct: 214 GPF--PSFLDQSPHKQVPSLGSNLWKEDTG-CLDWLESKEPRSVVYVNFGSITVMSAEQL 384
Query: 301 DEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSHR 360
E A L + + PFLW+IR + G E N E + IA WC Q +VL+H
Sbjct: 385 LEFAWGLANSKKPFLWIIRP-DLVIGGSVILSSEFVN--ETRDRSLIASWCPQEQVLNHP 555
Query: 361 SMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIV 420
S+G FLTHCGWNST ES+ + VPM+ +P + DQ TN + I + W+IG+ +D + +
Sbjct: 556 SIGVFLTHCGWNSTTESICAGVPMLCWPFFADQPTNCRYICNEWEIGMEIDTNAKRE--- 726
Query: 421 EGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKE 455
E E++ L V GEKG+++ + + K A+E
Sbjct: 727 ELEKLVNELMV----GEKGKKMGQKTMELKKKAEE 819
>TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, partial (46%)
Length = 993
Score = 140 bits (354), Expect = 9e-34
Identities = 87/203 (42%), Positives = 123/203 (59%), Gaps = 12/203 (5%)
Frame = +1
Query: 266 QAPSNDY--IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEK 323
QA N++ ++WLD + SVVYV FGS L +Q+ E+A AL D + PF+WVIRE K
Sbjct: 223 QASINEHHCLKWLDLQKSKSVVYVCFGSLCNLIPSQLVELALALEDTKRPFVWVIREGSK 402
Query: 324 FQDGEKEKEEELGNIEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRV 382
+Q+ EK EE G E + G I + W QV +LSH ++G FLTHCGWNST+E + + +
Sbjct: 403 YQELEKWISEE-GFEERTKGRGLIIRGWAPQVLILSHHAIGGFLTHCGWNSTLEGIGAGL 579
Query: 383 PMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHV-------NEDGI-VEGEEIRRGLTVVM- 433
PM+ +P + DQ N KL+ V KIGV V + V + G+ V+ E+I R + +VM
Sbjct: 580 PMITWPLFADQFLNEKLVTKVLKIGVSVGVEVPMKFGEEEKTGVLVKKEDINRAICMVMD 759
Query: 434 KNGEKGEELRRNAKKWKGLAKEA 456
+GE+ +E R A K +AK A
Sbjct: 760 DDGEESKERRERATKLSEMAKRA 828
>TC220150 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin
5-O-glucosyltransferase, partial (12%)
Length = 487
Score = 92.8 bits (229), Expect(2) = 2e-33
Identities = 52/80 (65%), Positives = 60/80 (75%), Gaps = 1/80 (1%)
Frame = +2
Query: 202 KEDFKELRG*ANTTILVNTFEALEPEAPRAVDKLNMIPIG-LLIPSAFLDRKDFAHDTSF 260
K F++L N ILVNTF+ LEP+A RAVDK MIPIG L IPSAFLD KD A DTS+
Sbjct: 254 KNSFRDLDDETNPIILVNTFQDLEPDALRAVDKFTMIPIGPLNIPSAFLDGKDPA-DTSY 430
Query: 261 GGDIIQAPSNDYIEWLDSKP 280
GGD+ A SNDY+EWLDS+P
Sbjct: 431 GGDLFDA-SNDYVEWLDSQP 487
Score = 68.2 bits (165), Expect(2) = 2e-33
Identities = 39/91 (42%), Positives = 55/91 (59%), Gaps = 2/91 (2%)
Frame = +1
Query: 118 HTLFVPWAPQVARELNLITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESSCSIEFSGL 177
+T+ +PWA +VAREL++ A LWI+ TV I Y YF +Y N +S +IE GL
Sbjct: 4 YTILLPWAAKVARELHIPGALLWIQAATVFDIYYYYF--HEYGDSFN-YKSDPTIELPGL 174
Query: 178 PFSLSPHDMPSYLISKGSY--VLPLFKEDFK 206
PFSL+ D+PS+L+ Y LP +E F+
Sbjct: 175 PFSLTARDVPSFLLPSNIYRFALPTLQEQFQ 267
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 134 bits (338), Expect = 7e-32
Identities = 104/329 (31%), Positives = 163/329 (48%), Gaps = 13/329 (3%)
Frame = +3
Query: 144 TTVMSILYNYFLGGDYIAKKNTVESSCSIEFSGLPFSLSPHDMPSYLISKGSYVLPLFKE 203
+T+ +LY + I +K+T + I+ GL +++ D P+ SY +F +
Sbjct: 135 STLALLLYYPTIHPTLIEEKDT-DQPLQIQIPGLS-TITADDFPNECKDPLSYACQVFLQ 308
Query: 204 DFKELRG*ANTTILVNTFEALEPEAPRAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGD 263
+ + G A I+VNTFEA+E EA RA+ + +P L G
Sbjct: 309 IAETMMGGAG--IIVNTFEAIEEEAIRALSEDATVPPPLFCV----------------GP 434
Query: 264 IIQAPSNDY----IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIR 319
+I AP + + WL+ +P SVV + FGS S+ Q+ EIA L FLWV+R
Sbjct: 435 VISAPYGEEDKGCLSWLNLQPSQSVVLLCFGSMGRFSRAQLKEIAIGLEKSEQRFLWVVR 614
Query: 320 EKEKFQDGEKEKEEEL--------GNIEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCG 370
+ G + EEL G +E +E G + + W Q +LSH S+G F+THCG
Sbjct: 615 TE---LGGADDSAEELSLDELLPEGFLERTKEKGMVVRDWAPQAAILSHDSVGGFVTHCG 785
Query: 371 WNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLT 430
WNS +E++ VPMVA+P + +Q N ++ K+ + V N+DG V E+ +
Sbjct: 786 WNSVLEAVCEGVPMVAWPLYAEQKMNRMVMVKEMKVALAV--KENKDGFVSSTELGDRVR 959
Query: 431 VVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
+M++ +KG+E+R+ K K A EA E
Sbjct: 960 ELMES-DKGKEIRQRIFKMKMSAAEAMAE 1043
>TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltransferase ,
partial (50%)
Length = 935
Score = 134 bits (337), Expect = 9e-32
Identities = 73/184 (39%), Positives = 109/184 (58%)
Frame = +2
Query: 273 IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKE 332
I+WLD PSSV+YV++GS V+S+ + E A L + PFLW+ R + + +
Sbjct: 89 IQWLDQWEPSSVIYVNYGSITVMSEDHLKEFAWGLANSNLPFLWIKRPDLVMGESTQLPQ 268
Query: 333 EELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTD 392
+ L +E+++ G I WC Q +VLSH S+G FLTHCGWNST+E + VPM+ +P + +
Sbjct: 269 DFL---DEVKDRGYITGWCPQEQVLSHPSVGVFLTHCGWNSTLEGISGGVPMIGWPFFAE 439
Query: 393 QNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGL 452
Q TN + I W IG + + +D V+ EE+ L M GE+G+E+R+ +WK
Sbjct: 440 QQTNCRYICTTWGIG----MDIKDD--VKREEVTT-LVKEMITGERGKEMRQKCLEWKKK 598
Query: 453 AKEA 456
A EA
Sbjct: 599 AIEA 610
>TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (35%)
Length = 1017
Score = 134 bits (336), Expect = 1e-31
Identities = 81/200 (40%), Positives = 114/200 (56%), Gaps = 9/200 (4%)
Frame = +2
Query: 269 SNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDG- 327
S + WLD +PP SV+YVSFGS LS+ Q++E+A L FLWV+R
Sbjct: 2 SGKCLSWLDKQPPCSVLYVSFGSGGTLSQNQINELASGLELSGQRFLWVLRAPSNSVSAA 181
Query: 328 --EKEKEEEL-----GNIEELEEVGKI-AKWCSQVEVLSHRSMGCFLTHCGWNSTMESLV 379
E KE+ L G +E +E G + A W QV+VL H S+G FL+HCGWNST+ES+
Sbjct: 182 YLEASKEDPLQFLPSGFLERTKEKGLVVASWAPQVQVLGHNSVGGFLSHCGWNSTLESVQ 361
Query: 380 SRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKG 439
VP++ +P + +Q NA ++ D K+ +R NEDGIVE EEI + + +M +GE+G
Sbjct: 362 EGVPLITWPLFAEQRMNAVMLTDGLKVALRPKF--NEDGIVEKEEIAKVIKCLM-DGEEG 532
Query: 440 EELRRNAKKWKGLAKEAGKE 459
+R K A A K+
Sbjct: 533 IGMRERMGNLKDSAASALKD 592
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 131 bits (330), Expect = 6e-31
Identities = 134/468 (28%), Positives = 209/468 (44%), Gaps = 24/468 (5%)
Frame = +1
Query: 16 IVSYPGQGQINPTLQFSKRLIAMGA--HVTIVTTLHMLRRMTTKTTIPGLSYTAFSDGFD 73
+V PG + P L+FSKRL+ + H+T ++K + L T S
Sbjct: 13 VVPSPGFTHLVPILEFSKRLLHLHPEFHITCFIPSVGSSPTSSKAYVQTLPPTITS---- 180
Query: 74 DGFIVDVATHFNLYTPELKRRGSEFITNLILFAENEGHPFTC-------LIHTLFVPWAP 126
+ T ++ P + E NL L E C L+ +F A
Sbjct: 181 --IFLPPITLDHVSDPSVLALQIELSVNLSLPYIREELKSLCSRAKVVALVVDVFANGAL 354
Query: 127 QVARELNLITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESSCSIEFSG-LPFSLSPHD 185
A+ELNL++ +++ + ++ LY Y D I + E I+ G +P
Sbjct: 355 NFAKELNLLSY-IYLPQSAMLLSLYFYSTKLDEILSSESRELQKPIDIPGCVPIHNKDLP 531
Query: 186 MPSYLISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALEPEAPRAVD-----KLNMIPI 240
+P + +S Y F E K + +NTF LE A RA++ K + P+
Sbjct: 532 LPFHDLSGLGY--KGFLERSKRFH--VPDGVFMNTFLELESGAIRALEEHVKGKPKLYPV 699
Query: 241 GLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQM 300
G +I + H+ + + WLD + P+SV+YVSFGS LS+ Q
Sbjct: 700 GPII-----QMESIGHENGV----------ECLTWLDKQEPNSVLYVSFGSGGTLSQEQF 834
Query: 301 DEIARALLDCRHPFLWVIREKEKFQDG------EKEKEEEL--GNIEELEEVGKIA-KWC 351
+E+A L FLWV+R K+ E L G +E ++ G + W
Sbjct: 835 NELAFGLELSGKKFLWVVRAPSGVVSAGYLCAETKDPLEFLPHGFLERTKKQGLVVPSWA 1014
Query: 352 SQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVD 411
Q++VL H + G FL+HCGWNS +ES+V VP++ +P + +Q+ NA +I D K+ +R
Sbjct: 1015PQIQVLGHSATGGFLSHCGWNSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKVALRP- 1191
Query: 412 LHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
VNE G+VE EEI + + +M + E E+R+ K A A KE
Sbjct: 1192-KVNESGLVEREEIAKVVRGLMGDKE-SLEIRKRMGLLKIAAANAIKE 1329
>TC230019 weakly similar to UP|Q6WFW1 (Q6WFW1) Glucosyltransferase-like
protein , partial (36%)
Length = 1202
Score = 129 bits (324), Expect = 3e-30
Identities = 87/257 (33%), Positives = 141/257 (54%), Gaps = 13/257 (5%)
Frame = +2
Query: 217 LVNTFEALEP---EAPRAVDKLNMIPIG-LLIPSAFLDRKDFAHDTSFGGDIIQAPSNDY 272
+ NT + +EP + R +L + P+G LL P++ +D K A S G + A
Sbjct: 326 ICNTVQEIEPLGLQLLRNYLQLPVWPVGPLLPPASLMDSKHRAGKES--GIALDA----C 487
Query: 273 IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQ-DGEKEK 331
++WLDSK SSV+Y+SFGS ++ +QM +A L + F+W+IR F +GE
Sbjct: 488 MQWLDSKDESSVLYISFGSQNTITASQMMALAEGLEESGRSFIWIIRPPFGFDINGEFIA 667
Query: 332 EEELGNIEELEEVGK----IAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAF 387
E EE K + KW Q+E+LSH S G FL+HCGWNS +ESL VPM+ +
Sbjct: 668 EWLPKGFEERMRDTKRGLLVNKWGPQLEILSHSSTGAFLSHCGWNSVLESLSYGVPMIGW 847
Query: 388 PQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAK 447
P +Q N K++ V ++GV ++L + ++ G+++++ + + M+ KG+E++ A
Sbjct: 848 PLAAEQTYNLKML--VEEMGVAIELTQTVETVISGKQVKKVIEIAMEQEGKGKEMKEKAN 1021
Query: 448 KWKGLAKEA----GKER 460
+ +EA GKE+
Sbjct: 1022EIAAHMREAITEKGKEK 1072
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 127 bits (318), Expect = 1e-29
Identities = 118/464 (25%), Positives = 209/464 (44%), Gaps = 26/464 (5%)
Frame = +1
Query: 15 LIVSYPGQGQINPTLQFSKRLIAMGAHVTIVTTLHMLRRMTTK--TTIPGLSYTAFSDGF 72
+++ +P QG +N L S+ +++ V V T+ +R++T + +I + + AF
Sbjct: 49 VLIPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFHAFE--- 219
Query: 73 DDGFIVDVATHFNLYTPELKRRGSEFITNLILFAENEGH---PFTCLIHTLFVPWAPQVA 129
F P ++F +L+ E H P L+H+L A +V
Sbjct: 220 --------VPSFVSPPPNPNNEETDFPAHLLPSFEASSHLREPVRKLLHSLSSQ-AKRVI 372
Query: 130 RELNLITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESSCSI-------EFSGLPFSLS 182
+ + A + + T M + NY T S+C+ + G P +
Sbjct: 373 VIHDSVMASV-AQDATNMPNVENY-----------TFHSTCTFGTAVFYWDKMGRPL-VD 513
Query: 183 PHDMPSYLISKGSYVLPLF-----KEDFKELRG*ANTTILVNTFEALEPEAPRAVDKLN- 236
+P +G + + DF+++ N + NT A+E +++
Sbjct: 514 GMLVPEIPSMEGCFTTDFMNFMIAQRDFRKV----NDGNIYNTSRAIEGAYIEWMERFTG 681
Query: 237 ---MIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYI 293
+ +G P AF ++KD + +EWLD + P+SV+YVSFG+
Sbjct: 682 GKKLWALGPFNPLAF-EKKD------------SKERHFCLEWLDKQDPNSVLYVSFGTTT 822
Query: 294 VLSKTQMDEIARALLDCRHPFLWVIREKEKFQ--DGEKEKEEELGNI--EELEEVGKIAK 349
+ Q+ +IA L + F+WV+R+ +K DG + K E N E +E +G + +
Sbjct: 823 TFKEEQIKKIATGLEQSKQKFIWVLRDADKGDIFDGSEAKWNEFSNEFEERVEGMGLVVR 1002
Query: 350 -WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGV 408
W Q+E+LSH S G F++HCGWNS +ES+ VP+ A+P +DQ N+ LI +V KIG+
Sbjct: 1003DWAPQLEILSHTSTGGFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGL 1182
Query: 409 RVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGL 452
V + +V + + +M+ E G+++R A + K +
Sbjct: 1183VVKNWAQRNALVSASNVENAVRRLMETKE-GDDMRERAVRLKNV 1311
>TC209756
Length = 1356
Score = 126 bits (316), Expect = 2e-29
Identities = 80/236 (33%), Positives = 123/236 (51%)
Frame = +1
Query: 217 LVNTFEALEPEAPRAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWL 276
L NT LEP A K +PIG L+ S + DT+ +EWL
Sbjct: 181 LCNTTYDLEPGAFSVSPKF--LPIGPLMESDNSKSAFWEEDTTC------------LEWL 318
Query: 277 DSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELG 336
D +PP SV+YVSFGS V+ Q E+A AL PF+WV+R ++ +
Sbjct: 319 DQQPPQSVIYVSFGSLAVMDPNQFKELALALDLLDKPFIWVVRPCNDNKENVNAYAHDFH 498
Query: 337 NIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTN 396
+ GKI W Q ++L+H ++ F++HCGWNST+E + + VP + +P TDQ +
Sbjct: 499 GSK-----GKIVGWAPQKKILNHPALASFISHCGWNSTLEGICAGVPFLCWPCATDQYLD 663
Query: 397 AKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGL 452
I DVWKIG+ +D +E+GI+ EEIR+ + ++ + E+++ + K K +
Sbjct: 664 KSYICDVWKIGLGLD--KDENGIISREEIRKKVDQLLVD----EDIKARSLKLKDM 813
>TC230868 weakly similar to UP|Q94AB5 (Q94AB5) AT3g46660/F12A12_180
(Glucosyltransferase-like protein), partial (38%)
Length = 765
Score = 125 bits (314), Expect = 4e-29
Identities = 70/178 (39%), Positives = 100/178 (55%)
Frame = +1
Query: 273 IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKE 332
+ WLD + +SVVYVSFGS +S+ + EIA L + + PFLWVIR E +
Sbjct: 10 MSWLDQQDRNSVVYVSFGSIAAISEAEFLEIAWGLANSKQPFLWVIRPG-LIHGSEWFEP 186
Query: 333 EELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTD 392
G +E L G I KW Q +VLSH ++G F TH GWNST+ES+ VPM+ P + D
Sbjct: 187 LPSGFLENLGGRGYIVKWAPQEQVLSHPAVGAFWTHNGWNSTLESICEGVPMICMPCFAD 366
Query: 393 QNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWK 450
Q NAK VW++GV++ ++ E+ + + +M G++G E+R NA K
Sbjct: 367 QKVNAKYASSVWRVGVQLQNKLDRG------EVEKTIKTLMV-GDEGNEIRENALNLK 519
>TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (59%)
Length = 1152
Score = 124 bits (311), Expect = 9e-29
Identities = 80/253 (31%), Positives = 133/253 (51%), Gaps = 12/253 (4%)
Frame = +2
Query: 216 ILVNTFEALEPEAPRAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEW 275
+++N+FE LEP K+ + + P + ++ KD G I + +I+W
Sbjct: 62 VVMNSFEELEPAYATGYKKIRGDKLWCIGPVSLIN-KDHLDKAQRGTASIDV--SQHIKW 232
Query: 276 LDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKE-KE-- 332
LD + P +V+Y GS L+ Q+ E+ AL + PF+WVIRE ++ EK KE
Sbjct: 233 LDCQKPGTVIYACLGSLCNLTTPQLKELGLALEASKRPFIWVIREGGHSEELEKWIKEYG 412
Query: 333 -EELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWT 391
EE N L I W Q+ +LSH ++G F+THCGWNST+E++ + VPM+ +P +
Sbjct: 413 FEERTNARSL----LIRGWAPQILILSHPAIGGFITHCGWNSTLEAICAGVPMLTWPLFA 580
Query: 392 DQNTNAKLIEDVWKIGVRVDLHVN-------EDGI-VEGEEIRRGLTVVMKNGEKGEELR 443
DQ N L+ V K+GV+V + + E G+ V+ +++ R + +M + E+ R
Sbjct: 581 DQFLNESLVVHVLKVGVKVGVEIPLTWGKEVEIGVQVKKKDVERAIAKLMDETSESEKRR 760
Query: 444 RNAKKWKGLAKEA 456
+ ++ +A A
Sbjct: 761 KRVRELAEMANRA 799
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,108,850
Number of Sequences: 63676
Number of extensions: 291032
Number of successful extensions: 1831
Number of sequences better than 10.0: 174
Number of HSP's better than 10.0 without gapping: 1707
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1733
length of query: 475
length of database: 12,639,632
effective HSP length: 101
effective length of query: 374
effective length of database: 6,208,356
effective search space: 2321925144
effective search space used: 2321925144
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0108.9


