

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.8
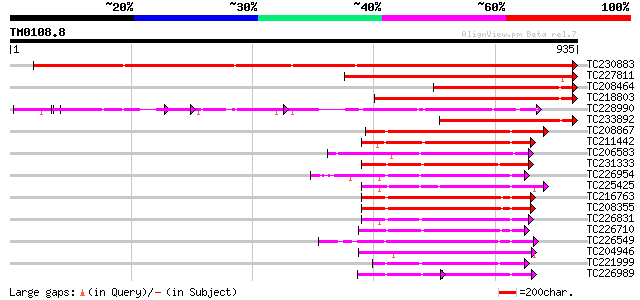
(935 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete 1368 0.0
TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , rece... 508 e-144
TC208464 homologue to UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RH... 422 e-118
TC218803 similar to UP|Q84P72 (Q84P72) Receptor-like protein kin... 406 e-113
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 298 1e-80
TC233892 similar to UP|Q84P72 (Q84P72) Receptor-like protein kin... 253 2e-67
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 239 5e-63
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 231 1e-60
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 229 4e-60
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 226 4e-59
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 219 3e-57
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 219 3e-57
TC216763 UP|Q9LKY3 (Q9LKY3) Pti1 kinase-like protein, complete 213 2e-55
TC208355 UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, complete 213 3e-55
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 213 4e-55
TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-l... 213 4e-55
TC226549 UP|Q84P43 (Q84P43) Protein kinase Pti1, complete 211 2e-54
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 210 3e-54
TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase... 206 4e-53
TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 120 9e-53
>TC230883 UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, complete
Length = 3053
Score = 1368 bits (3542), Expect = 0.0
Identities = 694/899 (77%), Positives = 768/899 (85%), Gaps = 3/899 (0%)
Frame = +1
Query: 40 MSKLLKSLSPPPSDWSSTTPFCQWDGIKCDSSNRVTTISLASRSLTGTLPSDLNSLSQLT 99
MS LKSL+PPPS WS TTPFCQW GI+CDSS+ VT+ISLAS+SLTGTLPSDLNSLSQL
Sbjct: 1 MSNFLKSLTPPPSGWSETTPFCQWKGIQCDSSSHVTSISLASQSLTGTLPSDLNSLSQLR 180
Query: 100 SLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSLSDNPNLSPW 159
+LSLQ+N+++G +PSL+NLS L+T + RNNF+SV +FA LT LQTLSL NP L PW
Sbjct: 181 TLSLQDNSLTGTLPSLSNLSFLQTVYFNRNNFSSVSPTAFASLTSLQTLSLGSNPALQPW 360
Query: 160 TLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVRLSYNNLTGALPNSL-AASA 218
+ PT+LT S+NLI L+L T LTG LP+ FDKFP LQ +RLSYNNLTG LP+S AA+
Sbjct: 361 SFPTDLTSSSNLIDLDLATVSLTGPLPD-IFDKFPSLQHLRLSYNNLTGNLPSSFSAANN 537
Query: 219 IENLWLNNQDNGLSGTIDVLSNMTQLAQVWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLT 278
+E LWLNNQ GLSGT+ VLSNM+ L Q WL+KNQFTG IPDLSQC+ L DLQLRDNQLT
Sbjct: 538 LETLWLNNQAAGLSGTLLVLSNMSALNQSWLNKNQFTGSIPDLSQCTALSDLQLRDNQLT 717
Query: 279 GPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDGINSFCKDTPGPCDARVMVLL 338
G VP SL L SL+ VSLDNNELQGP P FGKGV VTLDGINSFC DTPG CD RVMVLL
Sbjct: 718 GVVPASLTSLPSLKKVSLDNNELQGPVPVFGKGVNVTLDGINSFCLDTPGNCDPRVMVLL 897
Query: 339 HIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANLTDL 398
IA AFGYPI+ A SWKGNDPC GW++VVC +G KIITVN KQGLQGTISPAFANLTDL
Sbjct: 898 QIAEAFGYPIRLAESWKGNDPCDGWNYVVCAAG-KIITVNFEKQGLQGTISPAFANLTDL 1074
Query: 399 RSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTG 458
R+L+LNGNNL GSIP+SL TL QL+TL+VSDNNLSG VPKFPPKVKL+TAGN LLG+
Sbjct: 1075RTLFLNGNNLIGSIPDSLITLPQLQTLDVSDNNLSGLVPKFPPKVKLVTAGNALLGKPLS 1254
Query: 459 SGGGAGGKTTPSAGSTPEGSHGESGNG-SSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAK 517
GGG G TTPS GS+ GS GES G SS++PGWIAGIV+IVLFF+AVVLFVS KC+
Sbjct: 1255PGGGPSG-TTPS-GSSTGGSGGESSKGNSSVSPGWIAGIVVIVLFFIAVVLFVSWKCFVN 1428
Query: 518 RRHGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSI 577
+ GKFSRV NG G KLD + VSNGY G P ELQSQSSGD S+LH DG T SI
Sbjct: 1429KLQGKFSRVKGHENGKGGFKLDAVHVSNGYGGVPVELQSQSSGDRSDLHALDG--PTFSI 1602
Query: 578 LVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVL 637
VL+QVT NFSE+NILGRGGFGVVYKG+L DGTKIAVKRMESVAMGNKGL EF+AEI VL
Sbjct: 1603QVLQQVTNNFSEENILGRGGFGVVYKGQLHDGTKIAVKRMESVAMGNKGLKEFEAEIAVL 1782
Query: 638 SKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDV 697
SKVRHRHLVALLG+CING ERLLVYEYMPQGTLTQHLFEW+E GY PLTWKQRV +ALDV
Sbjct: 1783SKVRHRHLVALLGYCINGIERLLVYEYMPQGTLTQHLFEWQEQGYVPLTWKQRVVIALDV 1962
Query: 698 ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFG 757
ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFG
Sbjct: 1963ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFG 2142
Query: 758 YLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENI 817
YLAPEYAATGRVTTKVD+YAFG+VLMELITGR+ALDD++PDERSHLVTWFRRVLINKENI
Sbjct: 2143YLAPEYAATGRVTTKVDIYAFGIVLMELITGRKALDDTVPDERSHLVTWFRRVLINKENI 2322
Query: 818 PKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSR-H 876
PKAIDQTLNPDEETMESIYKV+ELAGHCTAREP QRPDMGHAVNVLVP+VEQWKP+S
Sbjct: 2323PKAIDQTLNPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLVPLVEQWKPSSHDE 2502
Query: 877 EDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
E+DG + MSLPQ L+RWQANEGTS+IFND+S+SQTQSSI+SK GFAD+FDS+DCR
Sbjct: 2503EEDGSGGDLQMSLPQALRRWQANEGTSSIFNDISISQTQSSISSKPVGFADTFDSMDCR 2679
>TC227811 similar to PIR|JQ1674|JQ1674 protein kinase TMK1 , receptor type
precursor - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (40%)
Length = 1436
Score = 508 bits (1308), Expect = e-144
Identities = 249/389 (64%), Positives = 311/389 (79%), Gaps = 6/389 (1%)
Frame = +2
Query: 553 ELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKI 612
E ++ + S++ + + GN +SI VL+ VT NFSE N+LG+GGFG VY+GEL DGT+I
Sbjct: 8 ETRTVPGSEASDIQMVEAGNMVISIQVLKNVTDNFSEKNVLGQGGFGTVYRGELHDGTRI 187
Query: 613 AVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQ 672
AVKRME A+ KG EF++EI VL+KVRHRHLV+LLG+C++GNE+LLVYEYMPQGTL++
Sbjct: 188 AVKRMECGAIAGKGAAEFKSEIAVLTKVRHRHLVSLLGYCLDGNEKLLVYEYMPQGTLSR 367
Query: 673 HLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKV 732
HLF+W E G PL W +R+T+ALDVARGVEYLH LA QSFIHRDLKPSNILLGDDMRAKV
Sbjct: 368 HLFDWPEEGLEPLEWNRRLTIALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKV 547
Query: 733 ADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRAL 792
ADFGLV+ AP+GK S+ETR+AGTFGYLAPEYA TGRVTTKVDV++FGV+LMELITGR+AL
Sbjct: 548 ADFGLVRLAPEGKASIETRIAGTFGYLAPEYAVTGRVTTKVDVFSFGVILMELITGRKAL 727
Query: 793 DDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQ 852
D++ P++ HLVTWFRR+ INK++ KAID T+ +EET+ SI+ V+ELAGHC AREP Q
Sbjct: 728 DETQPEDSMHLVTWFRRMSINKDSFRKAIDSTIELNEETLASIHTVAELAGHCGAREPYQ 907
Query: 853 RPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDM--- 909
RPDMGHAVNVL +VE WKP+ ++ +D + + MSLPQ L++WQA EG S + +
Sbjct: 908 RPDMGHAVNVLSSLVELWKPSDQNSEDIYGIDLDMSLPQALKKWQAYEGRSQMESSASSS 1087
Query: 910 ---SLSQTQSSINSKTYGFADSFDSLDCR 935
SL TQ+SI ++ YGFADSF S D R
Sbjct: 1088LLPSLDNTQTSIPTRPYGFADSFTSADGR 1174
>TC208464 homologue to UP|Q8LKR3 (Q8LKR3) Receptor-like kinase RHG4, partial
(26%)
Length = 893
Score = 422 bits (1086), Expect = e-118
Identities = 208/236 (88%), Positives = 224/236 (94%)
Frame = +2
Query: 700 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYL 759
GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYL
Sbjct: 2 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYL 181
Query: 760 APEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPK 819
APEYAATGRVTTKVDVYAFGVVLMELITGRRALDD++PDERSHLV+WFRRVLINKENIPK
Sbjct: 182 APEYAATGRVTTKVDVYAFGVVLMELITGRRALDDTVPDERSHLVSWFRRVLINKENIPK 361
Query: 820 AIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDD 879
AIDQTL+PDEETMESIYKV+ELAGHCTAREP QRPDMGHAVNVL P+VEQWKPT+ E++
Sbjct: 362 AIDQTLDPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLGPLVEQWKPTTHEEEE 541
Query: 880 GHDSEPHMSLPQVLQRWQANEGTSTIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
G+ + HMSLPQ L+RWQANEGTST+F DMS+SQTQSSI +K GFADSFDS+DCR
Sbjct: 542 GYGIDLHMSLPQALRRWQANEGTSTMF-DMSISQTQSSIPAKPSGFADSFDSMDCR 706
>TC218803 similar to UP|Q84P72 (Q84P72) Receptor-like protein kinase-like
protein (Fragment), partial (35%)
Length = 1266
Score = 406 bits (1043), Expect = e-113
Identities = 202/335 (60%), Positives = 255/335 (75%), Gaps = 1/335 (0%)
Frame = +1
Query: 602 YKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLV 661
++GEL DGTKIAVKRME+ + +K L+EFQ+EI VLSKVRHRHLV+LLG+ GNER+LV
Sbjct: 7 FRGELDDGTKIAVKRMEAGVISSKALDEFQSEIAVLSKVRHRHLVSLLGYSTEGNERILV 186
Query: 662 YEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSN 721
YEYMPQG L++HLF W+ PL+WK+R+ +ALDVARG+EYLH+LA QSFIHRDLKPSN
Sbjct: 187 YEYMPQGALSKHLFHWKSHDLEPLSWKRRLNIALDVARGMEYLHTLAHQSFIHRDLKPSN 366
Query: 722 ILLGDDMRAKVADFGLVKNAPDG-KYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGV 780
ILL DD +AKV+DFGLVK AP+G K SV TRLAGTFGYLAPEYA TG++TTK DV++FGV
Sbjct: 367 ILLADDFKAKVSDFGLVKLAPEGEKASVVTRLAGTFGYLAPEYAVTGKITTKADVFSFGV 546
Query: 781 VLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSE 840
VLMEL+TG ALD+ P+E +L WF + +K+ + AID L+ EET ES+ ++E
Sbjct: 547 VLMELLTGLMALDEDRPEESQYLAAWFWHIKSDKKKLMAAIDPALDVKEETFESVSIIAE 726
Query: 841 LAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQANE 900
LAGHCTAREP+QRPDMGHAVNVL P+VE+WKP ++ + + L Q+++ WQ E
Sbjct: 727 LAGHCTAREPSQRPDMGHAVNVLAPLVEKWKPFDDDTEEYSGIDYSLPLNQMVKGWQEAE 906
Query: 901 GTSTIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
G + M L ++SSI ++ GFADSF S D R
Sbjct: 907 GKDLSY--MDLEDSKSSIPARPTGFADSFTSADGR 1005
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 298 bits (762), Expect = 1e-80
Identities = 238/828 (28%), Positives = 380/828 (45%), Gaps = 36/828 (4%)
Frame = +1
Query: 85 TGTLPSDLNSLSQLTSLSLQNNAISGPIPS-LANLSALKTAFLGRNNFTSVPSASFAGLT 143
+G +P ++ +LS L L +SG IP+ L L L T FL N + + L
Sbjct: 757 SGGIPPEIGNLSNLVRLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLK 936
Query: 144 DLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVRLSY 203
L+++ LS+N + +P + NL L L +L G +PE F + P L+ ++L
Sbjct: 937 SLKSMDLSNN--MLSGEVPASFAELKNLTLLNLFRNKLHGAIPE-FVGELPALEVLQLWE 1107
Query: 204 NNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNM---TQLAQVWLHKNQFTGPIPD 260
NN TG++P +L + L ++ N ++GT+ NM +L + N GPIPD
Sbjct: 1108 NNFTGSIPQNLGNNGRLTL-VDLSSNKITGTLP--PNMCYGNRLQTLITLGNYLFGPIPD 1278
Query: 261 -LSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGK------GVK 313
L +C +L +++ +N L G +P L GL L V L +N L G FP G +
Sbjct: 1279 SLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQIS 1458
Query: 314 VTLDGINSFCKDTPGPCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRK 373
++ + ++ T G + +LL+ + G P Q ++
Sbjct: 1459 LSNNQLSGSLPSTIGNFTSMQKLLLN-----------GNEFTGRIPPQIGML------QQ 1587
Query: 374 IITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLS 433
+ ++ + G I+P + L + L+GN L+G IP +T++ L L +S N+L
Sbjct: 1588 LSKIDFSHNKFSGPIAPEISKCKLLTFIDLSGNELSGEIPNKITSMRILNYLNLSRNHLD 1767
Query: 434 GEVPKFPPKVKLLTAGNVLLGQTTGSGGGAG---------------------GKTTPSAG 472
G +P ++ LT+ + +G G G G
Sbjct: 1768 GSIPGNIASMQSLTSVDFSYNNFSGLVPGTGQFGYFNYTSFLGNPELCGPYLGPCKDGVA 1947
Query: 473 STPEGSHGESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNG 532
+ P H + SSL + G+++ + F +F +
Sbjct: 1948 NGPRQPHVKGPFSSSLKLLLVIGLLVCSILFAVAAIFKA--------------------- 2064
Query: 533 NGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNI 592
L+ S +L F + T+ V EDNI
Sbjct: 2065 -------------------RALKKASEARAWKLTAFQRLDFTVD-----DVLDCLKEDNI 2172
Query: 593 LGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHC 652
+G+GG G+VYKG + +G +AVKR+ +++ G+ + F AEI L ++RHRH+V LLG C
Sbjct: 2173 IGKGGAGIVYKGAMPNGDNVAVKRLPAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFC 2352
Query: 653 INGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSF 712
N LLVYEYMP G+L + + ++ G+ L W R +A++ A+G+ YLH
Sbjct: 2353 SNHETNLLVYEYMPNGSLGE-VLHGKKGGH--LHWDTRYKIAVEAAKGLCYLHHDCSPLI 2523
Query: 713 IHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GKYSVETRLAGTFGYLAPEYAATGRVTT 771
+HRD+K +NILL + A VADFGL K D G + +AG++GY+APEYA T +V
Sbjct: 2524 VHRDVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYTLKVDE 2703
Query: 772 KVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVL-INKENIPKAIDQTLN--PD 828
K DVY+FGVVL+EL+TGR+ + + + +V W R++ NKE + K +D L P
Sbjct: 2704 KSDVYSFGVVLLELVTGRKPVGEF--GDGVDIVQWVRKMTDSNKEGVLKVLDSRLPSVPL 2877
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRH 876
E M Y +A C + +RP M V +L + + P+S+H
Sbjct: 2878 HEVMHVFY----VAMLCVEEQAVERPTMREVVQILTELPK--PPSSKH 3003
Score = 160 bits (405), Expect = 2e-39
Identities = 149/472 (31%), Positives = 227/472 (47%), Gaps = 17/472 (3%)
Frame = +1
Query: 6 TLLSLSTLSLLFSITIIIAADDGPVFAA--TEDAAVMSKLLKSLSPPP----SDWSSTTP 59
+L+S+S L + ++ + AA +E A++S SL+ P S W+S+TP
Sbjct: 67 SLISISILRQKMRVLVLFFLFLHSLQAARISEYRALLSFKASSLTDDPTHALSSWNSSTP 246
Query: 60 FCQWDGIKCDSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIP-SLANL 118
FC W G+ CDS VT+++L S SL+GTL DL+ L L+ LSL +N SGPIP S + L
Sbjct: 247 FCSWFGLTCDSRRHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIPASFSAL 426
Query: 119 SALKTAFLGRNNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGT 178
SAL+ L N F + + L +L+ L L +N N++ LP + L L LG
Sbjct: 427 SALRFLNLSNNVFNATFPSQLNRLANLEVLDLYNN-NMT-GELPLSVAAMPLLRHLHLGG 600
Query: 179 ARLTGQLPESFFDKFPGLQSVRLSYNNLTGALPNSLA-ASAIENLWL---NNQDNGLSGT 234
+GQ+P + + LQ + LS N L G + L S++ L++ N G+
Sbjct: 601 NFFSGQIPPE-YGTWQHLQYLALSGNELAGTIAPELGNLSSLRELYIGYYNTYSGGIPPE 777
Query: 235 IDVLSNMTQLAQVWLHKNQFTGPIP-DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQN 293
I LSN+ +L + +G IP +L + NL L L+ N L+G + L L SL++
Sbjct: 778 IGNLSNLVRLDAAYC---GLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLKSLKS 948
Query: 294 VSLDNNELQGPFPA-FGKGVKVTLDGINSFCKDTPGPCDARVMVLLHIAGAFGYPIKFAG 352
+ L NN L G PA F + +TL +N F G V L + + F G
Sbjct: 949 MDLSNNMLSGEVPASFAELKNLTL--LNLFRNKLHGAIPEFVGELPALEVLQLWENNFTG 1122
Query: 353 SWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSI 412
S N + ++GR + V+L+ + GT+ P L++L GN L G I
Sbjct: 1123SIPQN---------LGNNGR-LTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPI 1272
Query: 413 PESLTTLTQLETLEVSDNNLSGEVPK----FPPKVKLLTAGNVLLGQTTGSG 460
P+SL L + + +N L+G +PK P ++ N+L GQ G
Sbjct: 1273PDSLGKCKSLNRIRMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDG 1428
Score = 94.7 bits (234), Expect = 2e-19
Identities = 70/240 (29%), Positives = 115/240 (47%), Gaps = 3/240 (1%)
Frame = +1
Query: 70 SSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIP-SLANLSALKTAFLGR 128
++ R+T + L+S +TGTLP ++ ++L +L N + GPIP SL +L +G
Sbjct: 1144 NNGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSLGKCKSLNRIRMGE 1323
Query: 129 NNFT-SVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPE 187
N S+P F GL L + L DN L P + + +T+L + L +L+G LP
Sbjct: 1324 NFLNGSIPKGLF-GLPKLTQVELQDN--LLTGQFPEDGSIATDLGQISLSNNQLSGSLP- 1491
Query: 188 SFFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNMTQLAQV 247
S F +Q + L+ N TG +P + + QL+++
Sbjct: 1492 STIGNFTSMQKLLLNGNEFTGRIPPQIGM------------------------LQQLSKI 1599
Query: 248 WLHKNQFTGPI-PDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFP 306
N+F+GPI P++S+C L + L N+L+G +PN + + L ++L N L G P
Sbjct: 1600 DFSHNKFSGPIAPEISKCKLLTFIDLSGNELSGEIPNKITSMRILNYLNLSRNHLDGSIP 1779
Score = 62.8 bits (151), Expect = 7e-10
Identities = 53/193 (27%), Positives = 81/193 (41%), Gaps = 2/193 (1%)
Frame = +1
Query: 73 RVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPS-LANLSALKTAFLGRNNF 131
++T + L LTG P D + + L +SL NN +SG +PS + N ++++ L N F
Sbjct: 1369 KLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQLSGSLPSTIGNFTSMQKLLLNGNEF 1548
Query: 132 TSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFD 191
T L L + S N P I E+ +L
Sbjct: 1549 TGRIPPQIGMLQQLSKIDFSHNKFSGP-------------IAPEISKCKL---------- 1659
Query: 192 KFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTI-DVLSNMTQLAQVWLH 250
L + LS N L+G +PN + + I N +LN N L G+I +++M L V
Sbjct: 1660 ----LTFIDLSGNELSGEIPNKITSMRILN-YLNLSRNHLDGSIPGNIASMQSLTSVDFS 1824
Query: 251 KNQFTGPIPDLSQ 263
N F+G +P Q
Sbjct: 1825 YNNFSGLVPGTGQ 1863
>TC233892 similar to UP|Q84P72 (Q84P72) Receptor-like protein kinase-like
protein (Fragment), partial (21%)
Length = 707
Score = 253 bits (647), Expect = 2e-67
Identities = 128/227 (56%), Positives = 164/227 (71%)
Frame = +2
Query: 709 QQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGR 768
+Q FIHRDLK SNILLGDD RAKV+DFGLVK APDGK SV TRLAGTFGYLAPEYA TG+
Sbjct: 5 EQIFIHRDLKSSNILLGDDFRAKVSDFGLVKLAPDGKKSVVTRLAGTFGYLAPEYAVTGK 184
Query: 769 VTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPD 828
VTTK DV++FGVVLMEL+TG ALD+ P+E +L +WF + +KE + AID L+
Sbjct: 185 VTTKADVFSFGVVLMELLTGLMALDEDRPEETQYLASWFWHIKSDKEKLMSAIDPALDIK 364
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMS 888
EE + + ++ELAGHC+AREPNQRPDM HAVNVL P+V++WKP ++ + +
Sbjct: 365 EEMFDVVSIIAELAGHCSAREPNQRPDMSHAVNVLSPLVQKWKPLDDETEEYSGIDYSLP 544
Query: 889 LPQVLQRWQANEGTSTIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
L Q+++ WQ EG + D L ++SSI ++ GFA+SF S+D R
Sbjct: 545 LNQMVKDWQETEGKDLSYVD--LQDSKSSIPARPTGFAESFTSVDGR 679
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 239 bits (609), Expect = 5e-63
Identities = 129/304 (42%), Positives = 187/304 (61%), Gaps = 3/304 (0%)
Frame = +1
Query: 588 SEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVA 647
S+ N++G GGFG+VY+G L DG K+A+K M+ G +G EF+ E+ +LS++ +L+A
Sbjct: 1 SKSNVIGHGGFGLVYRGVLNDGRKVAIKFMDQA--GKQGEEEFKVEVELLSRLHSPYLLA 174
Query: 648 LLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTP--LTWKQRVTVALDVARGVEYLH 705
LLG+C + N +LLVYE+M G L +HL+ TP L W+ R+ +AL+ A+G+EYLH
Sbjct: 175 LLGYCSDSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLH 354
Query: 706 SLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYS-VETRLAGTFGYLAPEYA 764
IHRD K SNILL AKV+DFGL K PD V TR+ GT GY+APEYA
Sbjct: 355 EHVSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYA 534
Query: 765 ATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQT 824
TG +TTK DVY++GVVL+EL+TGR +D P LV+W +L ++E + K +D +
Sbjct: 535 LTGHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPS 714
Query: 825 LNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSE 884
L + +M+ + +V+ +A C E + RP M V LVP+V+ + S+ +
Sbjct: 715 LE-GQYSMKEVVQVAAIAAMCVQPEADYRPLMADVVQSLVPLVKTQRSPSKVGSSSSFNS 891
Query: 885 PHMS 888
P +S
Sbjct: 892 PKLS 903
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 231 bits (588), Expect = 1e-60
Identities = 132/295 (44%), Positives = 183/295 (61%), Gaps = 8/295 (2%)
Frame = +2
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKG----ELQDGTK---IAVKRMESVAMGNKGLNEFQA 632
LR+ T +FS N+LG GGFG VYKG +L+ G K +AVKR++ G +G E+ A
Sbjct: 50 LREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRLD--LDGLQGHREWLA 223
Query: 633 EITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVT 692
EI L ++RH HLV L+G+C RLL+YEYMP+G+L LF + W R+
Sbjct: 224 EIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYSAA---MPWSTRMK 394
Query: 693 VALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYS-VETR 751
+AL A+G+ +LH A + I+RD K SNILL D AK++DFGL K+ P+G+ + V TR
Sbjct: 395 IALGAAKGLAFLHE-ADKPVIYRDFKASNILLDSDFTAKLSDFGLAKDGPEGEDTHVTTR 571
Query: 752 LAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVL 811
+ GT GY APEY TG +TTK DVY++GVVL+EL+TGRR +D S +E LV W R +L
Sbjct: 572 IMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNEGKSLVEWARPLL 751
Query: 812 INKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPM 866
+++ + ID+ L + M+ KV+ LA C + PN RP M + VL P+
Sbjct: 752 RDQKKVYNIIDRRLE-GQFPMKGAMKVAMLAFKCLSHHPNARPTMSDVIKVLEPL 913
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 229 bits (584), Expect = 4e-60
Identities = 136/352 (38%), Positives = 198/352 (55%), Gaps = 13/352 (3%)
Frame = +1
Query: 525 RVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSS---GDHSELHVFDGGN-STMSILVL 580
+ +P N N K +S G+ ++ S +S SE + N + ++ L
Sbjct: 88 KAESPYNTGFNSKY--VSTDGNDLGSTNDKVSANSIPQTPRSEGEILQSSNLKSFTLSEL 261
Query: 581 RQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLN--------EFQA 632
+ T NF D++LG GGFG V+KG + + + A K + + K LN E+ A
Sbjct: 262 KTATRNFRPDSVLGEGGFGSVFKGWIDENSLTATKPGTGIVIAVKRLNQDGIPGHREWLA 441
Query: 633 EITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVT 692
E+ L + H HLV L+G C+ RLLVYE+MP+G+ HLF R + PL+W R+
Sbjct: 442 EVNYLGQHFHSHLVRLIGFCLEDEHRLLVYEFMPRGSWENHLFR-RGSYFQPLSWSLRLK 618
Query: 693 VALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG-KYSVETR 751
VALD A+G+ +LHS A+ I+RD K SN+LL AK++DFGL K+ P G K V TR
Sbjct: 619 VALDAAKGLAFLHS-AEAKVIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPTGDKSHVSTR 795
Query: 752 LAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVL 811
+ GT+GY APEY ATG +T K DVY+FGVVL+E+++G+RA+D + P + +LV W + +
Sbjct: 796 VMGTYGYAAPEYLATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHNLVEWAKPFM 975
Query: 812 INKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
NK I + +D L T + YK++ LA C + E RP+M V L
Sbjct: 976 ANKRKIFRVLDTRLQGQYST-DDAYKLATLALRCLSIESKFRPNMDQVVTTL 1128
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 226 bits (576), Expect = 4e-59
Identities = 122/285 (42%), Positives = 177/285 (61%), Gaps = 1/285 (0%)
Frame = +3
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L+ TG FS+ N L GGFG V++G L DG IAVK+ + + +G EF +E+ VLS
Sbjct: 21 LQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLAS--TQGDKEFCSEVEVLSC 194
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVAR 699
+HR++V L+G C++ RLLVYEY+ G+L H++ ++ L W R +A+ AR
Sbjct: 195 AQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRKQ---NVLEWSARQKIAVGAAR 365
Query: 700 GVEYLHSLAQQS-FIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGY 758
G+ YLH + +HRD++P+NILL D A V DFGL + PDG VETR+ GTFGY
Sbjct: 366 GLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQPDGDMGVETRVIGTFGY 545
Query: 759 LAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIP 818
LAPEYA +G++T K DVY+FG+VL+EL+TGR+A+D + P + L W R L+ K+
Sbjct: 546 LAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEW-ARPLLEKQATY 722
Query: 819 KAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
K ID +L E +Y++ + + C R+P+ RP M + +L
Sbjct: 723 KLIDPSLRNCYVDQE-VYRMLKCSSLCIGRDPHLRPRMSQVLRML 854
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 219 bits (559), Expect = 3e-57
Identities = 139/377 (36%), Positives = 206/377 (53%), Gaps = 16/377 (4%)
Frame = +2
Query: 496 IVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQ 555
++I + + V+L + C++ R + +P N + SVS S +
Sbjct: 83 VIIEICLHLGVILLMGA-CWSNR-----IKAVSP----SNTGITSRSVSRSGHDISSNSR 232
Query: 556 SQSSG----DHSELHVFDGGN-STMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQD-- 608
S S+ SE + N + S LR T NF D++LG GGFG V+KG + +
Sbjct: 233 SSSASIPVTSRSEGEILQSSNLKSFSYHELRAATRNFRPDSVLGEGGFGSVFKGWIDEHS 412
Query: 609 --------GTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLL 660
G +AVK++ G +G E+ AEI L +++H +LV L+G+C RLL
Sbjct: 413 LAATKPGIGKIVAVKKLNQD--GLQGHREWLAEINYLGQLQHPNLVKLIGYCFEDEHRLL 586
Query: 661 VYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPS 720
VYE+MP+G++ HLF R + P +W R+ +AL A+G+ +LHS + I+RD K S
Sbjct: 587 VYEFMPKGSMENHLFR-RGSYFQPFSWSLRMKIALGAAKGLAFLHS-TEHKVIYRDFKTS 760
Query: 721 NILLGDDMRAKVADFGLVKNAPDG-KYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFG 779
NILL + AK++DFGL ++ P G K V TR+ GT GY APEY ATG +TTK DVY+FG
Sbjct: 761 NILLDTNYNAKLSDFGLARDGPTGDKSHVSTRVMGTRGYAAPEYLATGHLTTKSDVYSFG 940
Query: 780 VVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVS 839
VVL+E+I+GRRA+D + P +LV W + L NK + + +D L + + +
Sbjct: 941 VVLLEMISGRRAIDKNQPTGEHNLVEWAKPYLSNKRRVFRVMDPRLE-GQYSQNRAQAAA 1117
Query: 840 ELAGHCTAREPNQRPDM 856
LA C + EP RP+M
Sbjct: 1118ALAMQCFSVEPKCRPNM 1168
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 219 bits (559), Expect = 3e-57
Identities = 127/326 (38%), Positives = 186/326 (56%), Gaps = 17/326 (5%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQD--------GTKIAVKRMESVAMGNKGLNEFQ 631
L+ T NF D +LG GGFG V+KG L++ GT IAVK++ S ++ +GL E+Q
Sbjct: 127 LKAATRNFKADTVLGEGGFGKVFKGWLEEKATSKGGSGTVIAVKKLNSESL--QGLEEWQ 300
Query: 632 AEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRV 691
+E+ L ++ H +LV LLG+C+ +E LLVYE+M +G+L HLF R PL W R+
Sbjct: 301 SEVNFLGRLSHTNLVKLLGYCLEESELLLVYEFMQKGSLENHLFG-RGSAVQPLPWDIRL 477
Query: 692 TVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYS-VET 750
+A+ ARG+ +LH+ + I+RD K SNILL AK++DFGL K P S V T
Sbjct: 478 KIAIGAARGLAFLHT--SEKVIYRDFKASNILLDGSYNAKISDFGLAKLGPSASQSHVTT 651
Query: 751 RLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRV 810
R+ GT GY APEY ATG + K DVY FGVVL+E++TG RALD + P + L W +
Sbjct: 652 RVMGTHGYAAPEYVATGHLYVKSDVYGFGVVLVEILTGLRALDSNRPSGQHKLTEWVKPY 831
Query: 811 LINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL------- 863
L ++ + +D L + ++ +++++L+ C A EP RP M + L
Sbjct: 832 LHDRRKLKGIMDSRLE-GKFPSKAAFRIAQLSMKCLASEPKHRPSMKDVLENLERIQAAN 1008
Query: 864 -VPMVEQWKPTSRHEDDGHDSEPHMS 888
P+ +++ T GH + H S
Sbjct: 1009EKPVEPKFRSTHAASRQGHQAVHHRS 1086
>TC216763 UP|Q9LKY3 (Q9LKY3) Pti1 kinase-like protein, complete
Length = 1634
Score = 213 bits (543), Expect = 2e-55
Identities = 118/293 (40%), Positives = 183/293 (62%), Gaps = 5/293 (1%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L+ +T NF +G G +G VY+ L++G + +K+++S N+ EF ++++++S+
Sbjct: 373 LKSLTDNFGSKYFIGEGAYGKVYQATLKNGHAVVIKKLDS---SNQPEQEFLSQVSIVSR 543
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP---LTWKQRVTVAL 695
++H ++V L+ +C++G R L YEY P+G+L L + + G P L+W QRV +A+
Sbjct: 544 LKHENVVELVNYCVDGPFRALAYEYAPKGSLHDILHGRKGVKGAQPGPVLSWAQRVKIAV 723
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE-TRLAG 754
ARG+EYLH A+ IHR +K SNILL DD AKVADF L APD + TR+ G
Sbjct: 724 GAARGLEYLHEKAEIHIIHRYIKSSNILLFDDDVAKVADFDLSNQAPDAAARLHSTRVLG 903
Query: 755 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINK 814
TFGY APEYA TG++T+K DVY+FGV+L+EL+TGR+ +D +LP + LVTW L ++
Sbjct: 904 TFGYHAPEYAMTGQLTSKSDVYSFGVILLELLTGRKPVDHTLPRGQQSLVTWATPKL-SE 1080
Query: 815 ENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMV 867
+ + + +D L E +S+ K++ +A C E RP+M V L P++
Sbjct: 1081DKVKQCVDVRLK-GEYPSKSVAKMAAVAALCVQYEAEFRPNMSIIVKALQPLL 1236
>TC208355 UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, complete
Length = 1536
Score = 213 bits (542), Expect = 3e-55
Identities = 117/293 (39%), Positives = 184/293 (61%), Gaps = 5/293 (1%)
Frame = +2
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L+ +T NF +G G +G VY+ L++G + +K+++S N+ +EF ++++++S+
Sbjct: 383 LKPLTDNFGSKCFIGEGAYGKVYQATLKNGRAVVIKKLDS---SNQPEHEFLSQVSIVSR 553
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP---LTWKQRVTVAL 695
++H ++V L+ +C++G R L YEY P+G+L L + + G P L+W QRV +A+
Sbjct: 554 LKHENVVELVNYCVDGPFRALAYEYAPKGSLHDILHGRKGVKGAQPGPVLSWAQRVKIAV 733
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE-TRLAG 754
ARG+EYLH A+ IHR +K SNILL DD AK+ADF L APD + TR+ G
Sbjct: 734 GAARGLEYLHEKAEIHIIHRYIKSSNILLFDDDVAKIADFDLSNQAPDAAARLHSTRVLG 913
Query: 755 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINK 814
TFGY APEYA TG++T+K DVY+FGV+L+EL+TGR+ +D +LP + LVTW L ++
Sbjct: 914 TFGYHAPEYAMTGQLTSKSDVYSFGVILLELLTGRKPVDHTLPRGQQSLVTWATPKL-SE 1090
Query: 815 ENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMV 867
+ + + +D L E +S+ K++ +A C E RP+M V L P++
Sbjct: 1091DKVKQCVDVRLK-GEYPSKSVAKMAAVAALCVQYEAEFRPNMSIIVKALQPLL 1246
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 213 bits (541), Expect = 4e-55
Identities = 122/295 (41%), Positives = 175/295 (58%), Gaps = 11/295 (3%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQD----------GTKIAVKRMESVAMGNKGLNE 629
L+ T NF D++LG GGFG VYKG + + G +AVK+++ G +G E
Sbjct: 454 LKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPE--GLQGHKE 627
Query: 630 FQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQ 689
+ E+ L ++ H++LV L+G+C +G RLLVYE+M +G+L HLF G PL+W
Sbjct: 628 WLTEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFR---RGPQPLSWSV 798
Query: 690 RVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG-KYSV 748
R+ VA+ ARG+ +LH+ A+ I+RD K SNILL + AK++DFGL K P G + V
Sbjct: 799 RMKVAIGAARGLSFLHN-AKSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGDRTHV 975
Query: 749 ETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFR 808
T++ GT GY APEY ATGR+T K DVY+FGVVL+EL++GRRA+D S +LV W +
Sbjct: 976 STQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLVEWAK 1155
Query: 809 RVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
L +K + + +D L + + Y + LA C RE RP + + L
Sbjct: 1156PYLGDKRRLFRIMDTKLG-GQYPQKGAYMAATLALKCLNREAKGRPPITEVLQTL 1317
>TC226710 similar to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (66%)
Length = 1737
Score = 213 bits (541), Expect = 4e-55
Identities = 123/284 (43%), Positives = 171/284 (59%), Gaps = 2/284 (0%)
Frame = +3
Query: 576 SILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGT-KIAVKRMESVAMGNKGLNEFQAEI 634
S ++ T NF E +LG GGFG VYKGE+ GT K+A+KR ++ +G++EFQ EI
Sbjct: 402 SFAEIKAATNNFDEALLLGVGGFGKVYKGEIDGGTTKVAIKRGNPLS--EQGVHEFQTEI 575
Query: 635 TVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVA 694
+LSK+RHRHLV+L+G+C E +LVY+YM GTL +HL++ ++ P WKQR+ +
Sbjct: 576 EMLSKLRHRHLVSLIGYCEENTEMILVYDYMAYGTLREHLYKTQK---PPRPWKQRLEIC 746
Query: 695 LDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GKYSVETRLA 753
+ ARG+ YLH+ A+ + IHRD+K +NILL + AKV+DFGL K P V T +
Sbjct: 747 IGAARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVK 926
Query: 754 GTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLIN 813
G+FGYL PEY ++T K DVY+FGVVL E++ R AL+ +L E+ L W
Sbjct: 927 GSFGYLDPEYFRRQQLTDKSDVYSFGVVLFEVLCARPALNPTLAKEQVSLAEWAAH-CYQ 1103
Query: 814 KENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMG 857
K + ID L + E K +E A C A + RP MG
Sbjct: 1104KGILDSIIDPYLK-GKIAPECFKKFAETAMKCVADQGIDRPSMG 1232
>TC226549 UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1627
Score = 211 bits (536), Expect = 2e-54
Identities = 129/372 (34%), Positives = 210/372 (55%), Gaps = 8/372 (2%)
Frame = +3
Query: 509 FVSCKCYAKRRH--GKFSRVANPVN-GNGNVKLDVISVSNGYSGAPSELQSQSSGDHSEL 565
++ C C + + + + +P N G+GN K +S AP + ++Q + E
Sbjct: 207 WLCCTCQVEESYPSNENEHLKSPRNYGDGNQKGSKVS-------APVKPETQKAPPPIEA 365
Query: 566 HVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNK 625
+S+ L++ T NF ++G G +G VY L +G +AVK+++ V+ +
Sbjct: 366 -------PALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNNGKAVAVKKLD-VSSEPE 521
Query: 626 GLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP 684
NEF +++++S++++ + V L G+C+ GN R+L YE+ G+L L + + G P
Sbjct: 522 SNNEFLTQVSMVSRLKNGNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQP 701
Query: 685 ---LTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNA 741
L W QRV +A+D ARG+EYLH Q IHRD++ SN+L+ +D +AK+ADF L A
Sbjct: 702 GPTLDWIQRVRIAVDAARGLEYLHEKVQPPIIHRDIRSSNVLIFEDYKAKIADFNLSNQA 881
Query: 742 PDGKYSVE-TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDER 800
PD + TR+ GTFGY APEYA TG++T K DVY+FGVVL+EL+TGR+ +D ++P +
Sbjct: 882 PDMAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQ 1061
Query: 801 SHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAV 860
LVTW ++++ + + +D L E + + K+ +A C E RP+M V
Sbjct: 1062QSLVTW-ATPRLSEDKVKQCVDPKLK-GEYPPKGVAKLGAVAALCVQYEAEFRPNMSIVV 1235
Query: 861 NVLVPMVEQWKP 872
L P+++ P
Sbjct: 1236KALQPLLKSPAP 1271
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 210 bits (534), Expect = 3e-54
Identities = 126/308 (40%), Positives = 181/308 (57%), Gaps = 14/308 (4%)
Frame = +3
Query: 576 SILVLRQVTGNFSEDNILG-RGGFGVVYKGELQDGTKIAVKRMESVAMGNK--GLNEFQ- 631
S+ L T NF +D++LG G FG V+KG + + + A K V + K L+ FQ
Sbjct: 309 SLTELTAATRNFRKDSVLGGEGDFGSVFKGWIDNQSLAAAKPGTGVVVAVKRLSLDSFQG 488
Query: 632 -----AEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLT 686
AE+ L ++ H HLV L+G+C +RLLVYE+MP+G+L HLF R + PL+
Sbjct: 489 HKDRLAEVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLF-MRGSYFQPLS 665
Query: 687 WKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKY 746
W R+ VAL A+G+ +LHS A+ I+RD K SN+LL + AK+AD GL K+ P +
Sbjct: 666 WGLRLKVALGAAKGLAFLHS-AETKVIYRDFKTSNVLLDSNYNAKLADLGLAKDGPTREK 842
Query: 747 S-VETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVT 805
S TR+ GT+GY APEY ATG ++ K DV++FGVVL+E+++GRRA+D + P + +LV
Sbjct: 843 SHASTRVMGTYGYAAPEYLATGNLSAKSDVFSFGVVLLEMLSGRRAVDKNRPSGQHNLVE 1022
Query: 806 WFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL-- 863
W + L NK + + +D L E ++ KV+ L+ C A E RP M L
Sbjct: 1023WAKPYLSNKRKLLRVLDNRLEGQYE-LDEACKVATLSLRCLAIESKLRPTMDEVATDLEQ 1199
Query: 864 --VPMVEQ 869
VP V+Q
Sbjct: 1200LQVPHVKQ 1223
>TC221999 homologue to UP|Q9LK35 (Q9LK35) Receptor-protein kinase-like
protein, partial (38%)
Length = 1357
Score = 206 bits (524), Expect = 4e-53
Identities = 120/261 (45%), Positives = 159/261 (59%), Gaps = 1/261 (0%)
Frame = +1
Query: 598 FGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNE 657
FG VYKG L+DGT +AVKR +GL EF+ EI +LSK+RHRHLV+L+G+C +E
Sbjct: 1 FGRVYKGTLEDGTNVAVKRGNP--RSEQGLAEFRTEIEMLSKLRHRHLVSLIGYCDERSE 174
Query: 658 RLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDL 717
+LVYEYM G L HL+ PL+WKQR+ + + ARG+ YLH+ A QS IHRD+
Sbjct: 175 MILVYEYMANGPLRSHLYG---TDLPPLSWKQRLEICIGAARGLHYLHTGASQSIIHRDV 345
Query: 718 KPSNILLGDDMRAKVADFGLVKNAPD-GKYSVETRLAGTFGYLAPEYAATGRVTTKVDVY 776
K +NIL+ D+ AKVADFGL K P + V T + G+FGYL PEY ++T K DVY
Sbjct: 346 KTTNILVDDNFVAKVADFGLSKTGPALDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVY 525
Query: 777 AFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIY 836
+FGVVLME++ R AL+ LP E+ ++ W K + + +DQ L + S+
Sbjct: 526 SFGVVLMEVLCTRPALNPVLPREQVNIAEW-AMSWQKKGMLDQIMDQNL-VGKVNPASLK 699
Query: 837 KVSELAGHCTAREPNQRPDMG 857
K E A C A RP MG
Sbjct: 700 KFGETAEKCLAEYGVDRPSMG 762
>TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (86%)
Length = 1730
Score = 120 bits (302), Expect(2) = 9e-53
Identities = 67/157 (42%), Positives = 89/157 (56%), Gaps = 1/157 (0%)
Frame = +3
Query: 714 HRDLKPSNILLGDDMRAKVADFGLVKNAPDG-KYSVETRLAGTFGYLAPEYAATGRVTTK 772
+RD K SNILL + K++DFGL K P G K V TR+ GT+GY APEYA TG++T K
Sbjct: 657 NRDCKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVK 836
Query: 773 VDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETM 832
DVY+FGVV +ELITGR+A+D + P +LVTW R + ++ K D L M
Sbjct: 837 SDVYSFGVVFLELITGRKAIDSTQPQGEQNLVTWARPLFNDRRKFSKLADPRLQ-GRFPM 1013
Query: 833 ESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQ 869
+Y+ +A C RP +G V L + Q
Sbjct: 1014RGLYQALAVASMCIQESAATRPLIGDVVTALSYLANQ 1124
Score = 105 bits (263), Expect(2) = 9e-53
Identities = 59/146 (40%), Positives = 88/146 (59%), Gaps = 1/146 (0%)
Frame = +2
Query: 574 TMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKI-AVKRMESVAMGNKGLNEFQA 632
T + L T NF ++ +G GGFG VYKG L+ +I AVK+++ G +G EF
Sbjct: 242 TFTFRELAAATKNFRPESFVGEGGFGRVYKGRLETTAQIVAVKQLDK--NGLQGNREFLV 415
Query: 633 EITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVT 692
E+ +LS + H +LV L+G+C +G++RLLVYE+MP G+L HL + PL W R+
Sbjct: 416 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPP-DKEPLDWNTRMK 592
Query: 693 VALDVARGVEYLHSLAQQSFIHRDLK 718
+A+ A+G+EYLH A I + L+
Sbjct: 593 IAVGAAKGLEYLHDKANPPGIQQRLQ 670
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,414,294
Number of Sequences: 63676
Number of extensions: 577480
Number of successful extensions: 6314
Number of sequences better than 10.0: 1171
Number of HSP's better than 10.0 without gapping: 4594
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5254
length of query: 935
length of database: 12,639,632
effective HSP length: 106
effective length of query: 829
effective length of database: 5,889,976
effective search space: 4882790104
effective search space used: 4882790104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0108.8


