

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.16
(709 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
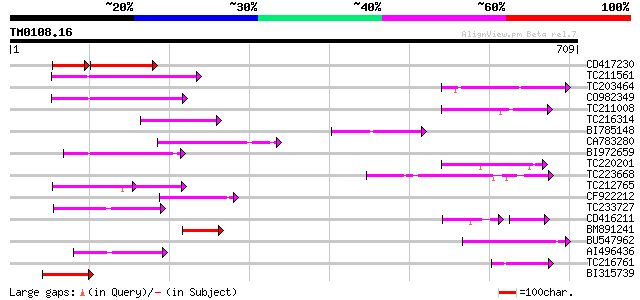
Score E
Sequences producing significant alignments: (bits) Value
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 65 2e-19
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 88 1e-17
TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leish... 79 5e-15
CO982349 79 9e-15
TC211008 75 8e-14
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 75 8e-14
BI785148 homologue to GP|25166165|dbj murF {Wigglesworthia brevi... 75 1e-13
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 71 1e-12
BI972659 68 1e-11
TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance ... 67 3e-11
TC223668 66 6e-11
TC212765 62 7e-10
CF922212 62 1e-09
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 58 1e-08
CD416211 41 2e-08
BM891241 55 1e-07
BU547962 54 2e-07
AI496436 53 4e-07
TC216761 similar to UP|Q9FQE4 (Q9FQE4) Glutathione S-transferase... 51 2e-06
BI315739 51 2e-06
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 65.1 bits (157), Expect(2) = 2e-19
Identities = 30/83 (36%), Positives = 52/83 (62%)
Frame = -2
Query: 102 SKGVHPSIRDDIRGIAPIPLVNDLGKYLGFPLSGGRVSRGRFNFLLENINRKMAAWKTNL 161
SK V+ + +DI + +DLGKYLG P+ + F+F+++ +N++ + WK+NL
Sbjct: 321 SKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRFSRWKSNL 142
Query: 162 LNLAGRACLAKSVIAAMPTYTMQ 184
L++ GR L K V+ A+P++ MQ
Sbjct: 141 LSMVGRLTLTK*VV*ALPSHIMQ 73
Score = 49.3 bits (116), Expect(2) = 2e-19
Identities = 23/46 (50%), Positives = 33/46 (71%)
Frame = -3
Query: 54 VSHLLFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKA 99
+SHL F DD++LF +A+ QV ++ TL F SSG KVN+DK+K+
Sbjct: 467 ISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTKS 330
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 87.8 bits (216), Expect = 1e-17
Identities = 50/188 (26%), Positives = 90/188 (47%)
Frame = +1
Query: 53 PVSHLLFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVHPSIRDD 112
P+S L +ADD + F +A+ V+ + LR F S L++N KS GV + +
Sbjct: 376 PISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKS-CFGVFGVTDQWKQE 552
Query: 113 IRGIAPIPLVNDLGKYLGFPLSGGRVSRGRFNFLLENINRKMAAWKTNLLNLAGRACLAK 172
L+ YLG P+ ++ ++ RK++ WK ++ GR L K
Sbjct: 553 AANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGGRVTLIK 732
Query: 173 SVIAAMPTYTMQVFWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDKSHGGLG 232
SV+ ++P Y F +P+S++ + + R+F+W G Q+ + + W + K GG+
Sbjct: 733 SVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPKERGGI* 912
Query: 233 MKDMSDHN 240
+ ++HN
Sbjct: 913 KRLPNNHN 936
>TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leishmania
major;} , partial (8%)
Length = 1386
Score = 79.3 bits (194), Expect = 5e-15
Identities = 57/168 (33%), Positives = 87/168 (50%), Gaps = 6/168 (3%)
Frame = +2
Query: 540 WKPPPNQCMKMNVDGS---FRSDEGLMGTGGALRDSSGSWVTGFMAHYDNGNAFI-AEML 595
WK P +K+NVDGS ++S G GG LRD+S W+ GF + A E+
Sbjct: 488 WKKPEIGWVKLNVDGSRDPYKSSSA--GCGGVLRDASAKWLRGFAKKLNPTYAVHQTELE 661
Query: 596 ALRDGLKIAWEQGCRRLICESDCLELVRNLASVDWVSLHSHGHILSEIRTMLR-WSWRID 654
A+ GLK+A E ++LI ESD +V + + + +G ++ IRT R + W +
Sbjct: 662 AILTGLKVASEMNVKKLIVESDSDSVVSMVENGVKPNHPDYG-VVELIRTKRRRFDWEVR 838
Query: 655 IAWIDREGNRVADWLAKRGASIALSEV-QELVEPPSELQILLLKDSLG 701
+ + NRVAD LA +A +V +E + PP+ LLL+D +G
Sbjct: 839 FVSVSNKANRVADRLANDARKLACDDVCEEYLHPPANCNQLLLEDEIG 982
>CO982349
Length = 795
Score = 78.6 bits (192), Expect = 9e-15
Identities = 47/170 (27%), Positives = 79/170 (45%)
Frame = +3
Query: 53 PVSHLLFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVHPSIRDD 112
PVS L +ADD + F +A+ ++++ LR F +SGLK+N +S+ + +
Sbjct: 279 PVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESR-FGAIWKPDHWCKE 455
Query: 113 IRGIAPIPLVNDLGKYLGFPLSGGRVSRGRFNFLLENINRKMAAWKTNLLNLAGRACLAK 172
+++ YLG P+ R + ++ K+A WK ++L GR L
Sbjct: 456 AAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHISLGGRVTLIN 635
Query: 173 SVIAAMPTYTMQVFWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTV 222
+++ A+P Y F P ++ + R F+W GS QR V W TV
Sbjct: 636 AILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRRIAWVKWETV 785
>TC211008
Length = 516
Score = 75.5 bits (184), Expect = 8e-14
Identities = 50/146 (34%), Positives = 80/146 (54%), Gaps = 7/146 (4%)
Frame = +1
Query: 540 WKPPPNQCMKMNVDGSFRSDE--GLMGT--GGALRDSSGSWVTGFMAHYDNGNAFIAEML 595
W+ P N + +N DGS + G G+ GG +RDSSG ++ GF + N + +AE+
Sbjct: 64 WRCPXNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAELW 243
Query: 596 ALRDGLKIAWEQGCRRL---ICESDCLELVRNLASVDWVSLHSHGHILSEIRTMLRWSWR 652
+ GLK+AW+ GC+++ I + L LVR+ V+ ++SEI ++R W
Sbjct: 244 GVVHGLKLAWDLGCKKVKVDIDSGNALGLVRH----GPVANDPAFALVSEINELVRKEWL 411
Query: 653 IDIAWIDREGNRVADWLAKRGASIAL 678
++ + + RE NR AD LA G S +L
Sbjct: 412 VEFSHVFRESNRAADKLAHLGHSHSL 489
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 75.5 bits (184), Expect = 8e-14
Identities = 34/101 (33%), Positives = 55/101 (53%)
Frame = +2
Query: 164 LAGRACLAKSVIAAMPTYTMQVFWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVV 223
+ GR L KSV++A+P + F +P+ I+ + R F+W V W+ +
Sbjct: 2 MGGRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADIC 181
Query: 224 RDKSHGGLGMKDMSDHNTALLGKAVWLLLKNSNKLWVQVMQ 264
K GGLG+KD+S+ N AL G+ +W L N N+LW ++ +
Sbjct: 182 NPKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARLAE 304
>BI785148 homologue to GP|25166165|dbj murF {Wigglesworthia brevipalpis},
partial (2%)
Length = 422
Score = 75.1 bits (183), Expect = 1e-13
Identities = 42/120 (35%), Positives = 58/120 (48%), Gaps = 1/120 (0%)
Frame = -2
Query: 403 WLCLHDALPVNSKRFHCHLANSEACSRCSYIREDGIHSLRDCTHSRELWSRMGAGRWNNF 462
WL LH++L NS RFH +L NS A CS RED +H LRD H++E+W +G G +
Sbjct: 394 WLILHNSLSKNSTRFHRNLVNSSAFPHCSNPREDSLHCLRDFPHAKEIW--LGFGFAPHL 221
Query: 463 WTLNLS-DWITLHARSDHAVKFLAGLWGIWKWRCNMVMDTAPWTIDTAWRKIAHDHDDFL 521
+L L + F +W + KW N + D W I + I HDD +
Sbjct: 220 SSLPLM*SGLKSQLNGSKVFLFTTIVWWV*KWGNNCIFDDEKWDIQRVVQSICFYHDDLV 41
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 71.2 bits (173), Expect = 1e-12
Identities = 49/155 (31%), Positives = 68/155 (43%)
Frame = +2
Query: 186 FWLPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDKSHGGLGMKDMSDHNTALLG 245
F LP II ++ R F+W + R VNW TV K+ GGLG+KD+ NT LLG
Sbjct: 11 FSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTLLG 190
Query: 246 KAVWLLLKNSNKLWVQVMQHKYLRDHTILNAPHRASSSAVWKGILRARDRLRGGFKFRLG 305
K W L + W +V+Q KY + + SA WK +++ + R +
Sbjct: 191 KWRWDLFYIQQEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQR-----NIP 355
Query: 306 TGNSSIWYNDWSGHGNLCEHLPFVHITDTQLHLRD 340
+IW E L T+T L LRD
Sbjct: 356 LKRETIWKVGGGDRIKFWEDL----WTNTDLSLRD 448
>BI972659
Length = 453
Score = 68.2 bits (165), Expect = 1e-11
Identities = 42/152 (27%), Positives = 73/152 (47%)
Frame = +1
Query: 68 QASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVHPSIRDDIRGIAPIPLVNDLGK 127
+AS + ++ LR F SGL++N KS+ G P+ D + ++
Sbjct: 7 EASWDNIIVLKSMLRGFEMVSGLRINYAKSQ-FGIVGFQPNWAHDAAQLLNCRQLDIPFH 183
Query: 128 YLGFPLSGGRVSRGRFNFLLENINRKMAAWKTNLLNLAGRACLAKSVIAAMPTYTMQVFW 187
YLG P++ SR + L+ K++ W L++ GR L KSV+ A+P Y + F
Sbjct: 184 YLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFK 363
Query: 188 LPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNW 219
+P+ I+ + R+F+W G + N ++W
Sbjct: 364 IPQRIVDKLVSLQRTFMW---GGNQHHNRISW 450
>TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance receptor
PtdSerR (Phosphatidylserine receptor), partial (5%)
Length = 1185
Score = 67.0 bits (162), Expect = 3e-11
Identities = 48/143 (33%), Positives = 72/143 (49%), Gaps = 10/143 (6%)
Frame = +1
Query: 540 WKPPPNQCMKMNVDGS-FRSDEGLMGTGGALRDSSGSWVTGFMAHYDNG---NAFIAEML 595
WK P + +K+NVDGS + G GG +RD G+W GF D A E+
Sbjct: 337 WKKPESGWVKLNVDGSRIHEEPASAGCGGVIRDEWGTWCVGFDQKLDPNICRQAHYTELQ 516
Query: 596 ALRDGLKIAWEQ--GCRRLICESDCLELVRNLASVDWVSLHSHGH-ILSEIRTMLR---W 649
A+ GLK+A E +L+ ESD V + S H + ++ +I +L+ W
Sbjct: 517 AILTGLKVAREDMINVEKLVVESDSEPAVNMVKSRLGYKYHRPEYKVVQDINRLLKDPKW 696
Query: 650 SWRIDIAWIDREGNRVADWLAKR 672
R+D ++ R+ NRVA++LAKR
Sbjct: 697 LARLD--YVPRKANRVANYLAKR 759
>TC223668
Length = 866
Score = 65.9 bits (159), Expect = 6e-11
Identities = 64/244 (26%), Positives = 103/244 (41%), Gaps = 11/244 (4%)
Frame = -3
Query: 447 SRELWSRMGAGRWNNFWTLNL-SDWITLHARSDHAVKFLAGLWGIWKWRCN-MVMDTAPW 504
+RELW R+ G + NL + + + +D A K L W WKW CN ++ W
Sbjct: 852 TRELWLRVSNGNGSLLSHTNLFAXXXAVLSNNDCAYKVLEAWWA-WKW-CN*VIFSPLIW 679
Query: 505 TIDTAWRKIAHDHDDFLKFMQPQLSHDDSFWLSVVWKPPPNQC--MKMNVDGSFRSDEGL 562
+ ++ I H +S D + W PP +K+NVDGS + +
Sbjct: 678 S----FQFIKKIHC*GYCLQSSLVSKDLVSSKRLTWTSPPPLLNEVKLNVDGSGHYEPHV 511
Query: 563 MGTGGALRDSSGSWVTGFMAHYDNGNAFIAEMLALRDGLKI----AWEQGCRRLICESDC 618
M GG +RD G W+ F H GN ++E+ AL L+ W C
Sbjct: 510 MEVGGLIRDGVGRWLFRFARHCGLGNPLLSELFALEASLRSHIV*TW----------WSC 361
Query: 619 L---ELVRNLASVDWVSLHSHGHILSEIRTMLRWSWRIDIAWIDREGNRVADWLAKRGAS 675
L ++ R + + +HS IR+ ++ W + ++ + N V D+LA+ GA
Sbjct: 360 LLIGQMSR*ACMLMQILIHS-------IRSFMKRDWSFSLCHVNTKVNSVVDFLAEWGAR 202
Query: 676 IALS 679
+ S
Sbjct: 201 MTKS 190
>TC212765
Length = 637
Score = 62.4 bits (150), Expect = 7e-10
Identities = 30/68 (44%), Positives = 40/68 (58%)
Frame = +2
Query: 154 MAAWKTNLLNLAGRACLAKSVIAAMPTYTMQVFWLPRSIIHHIDRAMRSFIWSKGSGQRG 213
+A+WK LLN A + CL V+ A P Y M+ WLP+SI ID+A R IW+K
Sbjct: 416 LASWKGKLLNQASKHCLT*LVLHAFPIYPMEASWLPQSIFQAIDKASRVTIWNKVGASLS 595
Query: 214 WNLVNWST 221
W+ V+ ST
Sbjct: 596 WHSVS*ST 619
Score = 53.1 bits (126), Expect = 4e-07
Identities = 39/121 (32%), Positives = 54/121 (44%), Gaps = 15/121 (12%)
Frame = +3
Query: 54 VSHLLFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAISSKGVHPSIRDDI 113
V +L+ DD+ LF A L + TL +FF + GLK+N+DKS S + P +RD I
Sbjct: 120 VPYLMLVDDIFLFINAIGD*AYLASLTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLI 299
Query: 114 RGIAPIPLVNDLGKYLGFPLSGGRVS---------------RGRFNFLLENINRKMAAWK 158
I LGKYL GR RGR +FL + +N +W
Sbjct: 300 SNTLNIVHAPSLGKYLCLKFIHGRSKIVDFLLVERIHNPWPRGRGSFLTKQVNIV*RSWS 479
Query: 159 T 159
+
Sbjct: 480 S 482
>CF922212
Length = 445
Score = 61.6 bits (148), Expect = 1e-09
Identities = 33/99 (33%), Positives = 49/99 (49%)
Frame = -2
Query: 188 LPRSIIHHIDRAMRSFIWSKGSGQRGWNLVNWSTVVRDKSHGGLGMKDMSDHNTALLGKA 247
+P ++ + R + F+W G GQ+ VNW +V K GLG +D+ N ALLGK
Sbjct: 441 VPNKVLDKLVRLQQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKR 262
Query: 248 VWLLLKNSNKLWVQVMQHKYLRDHTILNAPHRASSSAVW 286
W L + +L +V+ KY R L+ R S + W
Sbjct: 261 RWNLFHHQGELGARVLDSKYKRWRN-LDEERRVKSESFW 148
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 58.2 bits (139), Expect = 1e-08
Identities = 41/146 (28%), Positives = 71/146 (48%), Gaps = 5/146 (3%)
Frame = -2
Query: 55 SHLLFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAI--SSKGVHPSIRDD 112
SH++ ADD+++FC+ ++ V + + + + S G ++ KSK S G + D
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGSIPGQRLRVISD 272
Query: 113 IRGI--APIPLVNDLGKYLGFPLSGGRVSRGRFNFLLENINRKMAAWKTNLLNLAGRACL 170
+ + IP Y G P+ G+ +R + + I K+A+WK ++L++ GR L
Sbjct: 271 LLQYFHSDIPF-----NYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQL 107
Query: 171 AKSVIAAMPTYTMQVF-WLPRSIIHH 195
SVI M Y+ V+ W S H
Sbjct: 106 VNSVIHGMLLYSFSVYAWQVLSFERH 29
>CD416211
Length = 551
Score = 41.2 bits (95), Expect(2) = 2e-08
Identities = 22/51 (43%), Positives = 29/51 (56%)
Frame = -1
Query: 625 LASVDWVSLHSHGHILSEIRTMLRWSWRIDIAWIDREGNRVADWLAKRGAS 675
L SV S H ++ +IR ++ W + REG+RVADWLAK GAS
Sbjct: 263 LPSVFSWSTHPLAPLVGDIRGLVSAQWSVTFNHTLREGHRVADWLAKEGAS 111
Score = 35.8 bits (81), Expect(2) = 2e-08
Identities = 24/79 (30%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Frame = -3
Query: 542 PPPNQCMKMNVDGSFRSDEGLMGTGGALRDSSG---SWVTGFMAHYDNGNAFIAEMLALR 598
PP +K+N DGS + G+ G GG R+ G SW G YD +
Sbjct: 474 PPDPNTIKINTDGSSFGNPGISGYGGVFRNEHGHVASWFLGQQWVYD*HKCGV------- 316
Query: 599 DGLKIAWEQGCRRLICESD 617
+ + ++G + +ICESD
Sbjct: 315 --VSNSKQKG*QSVICESD 265
>BM891241
Length = 407
Score = 55.1 bits (131), Expect = 1e-07
Identities = 23/51 (45%), Positives = 32/51 (62%)
Frame = -2
Query: 217 VNWSTVVRDKSHGGLGMKDMSDHNTALLGKAVWLLLKNSNKLWVQVMQHKY 267
V W V K+ GGLG+KD+S N ALLGK +W L + +LW +++ KY
Sbjct: 373 VKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINSKY 221
>BU547962
Length = 591
Score = 53.9 bits (128), Expect = 2e-07
Identities = 39/135 (28%), Positives = 61/135 (44%)
Frame = -2
Query: 567 GALRDSSGSWVTGFMAHYDNGNAFIAEMLALRDGLKIAWEQGCRRLICESDCLELVRNLA 626
G +R+ + ++ + N+ IAE+ ALR GL++ E G + E D LV +
Sbjct: 590 GVVRNHNAEFLXXYAESIGQANSTIAELTALRKGLELVLENGWNDIWLEGDAKTLVEIIV 411
Query: 627 SVDWVSLHSHGHILSEIRTMLRWSWRIDIAWIDREGNRVADWLAKRGASIALSEVQELVE 686
V ++ I T+L ++ I REGNR AD A+ G + +
Sbjct: 410 KRRKVRCTEVQRHINHINTILPEFNNFFVSHIYREGNRAADKFAQMGHHLDQPRIWR--H 237
Query: 687 PPSELQILLLKDSLG 701
PP EL LL+D+ G
Sbjct: 236 PPDELLAQLLEDAQG 192
>AI496436
Length = 414
Score = 53.1 bits (126), Expect = 4e-07
Identities = 35/121 (28%), Positives = 54/121 (43%), Gaps = 4/121 (3%)
Frame = +1
Query: 81 LRDFFASSGLKVNIDKSK--AISSKGVHPSIRDDI--RGIAPIPLVNDLGKYLGFPLSGG 136
LR F SSGLK+N KS+ I + D + +P V YLG P+
Sbjct: 7 LRCFELSSGLKINFAKSQFGTICKSDIWRKEAADYLNSSVLSMPFV-----YLGIPIGAN 171
Query: 137 RVSRGRFNFLLENINRKMAAWKTNLLNLAGRACLAKSVIAAMPTYTMQVFWLPRSIIHHI 196
+ ++ RK+A+WK ++ GR L SV+ A+P Y + F +P ++
Sbjct: 172 SRHSDVWEPVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKVVDKA 351
Query: 197 D 197
D
Sbjct: 352 D 354
>TC216761 similar to UP|Q9FQE4 (Q9FQE4) Glutathione S-transferase GST 14
(Fragment) , complete
Length = 1353
Score = 51.2 bits (121), Expect = 2e-06
Identities = 29/77 (37%), Positives = 39/77 (49%)
Frame = +1
Query: 603 IAWEQGCRRLICESDCLELVRNLASVDWVSLHSHGHILSEIRTMLRWSWRIDIAWIDREG 662
IAW +G R LICESD + L S H +++ IR+ + W + REG
Sbjct: 1018 IAWTRGIRNLICESDS-SMALQLISTGVYITHPRAALVTTIRSFMDKDWCLSFQHTLREG 1194
Query: 663 NRVADWLAKRGASIALS 679
N VAD LAK+G + S
Sbjct: 1195 NFVADGLAKKGVHVTAS 1245
>BI315739
Length = 442
Score = 51.2 bits (121), Expect = 2e-06
Identities = 28/65 (43%), Positives = 42/65 (64%), Gaps = 1/65 (1%)
Frame = +2
Query: 42 LEAY*GFSPWP-PVSHLLFADDVLLFCQASTTQVQLVADTLRDFFASSGLKVNIDKSKAI 100
L +* F WP +SHL F D+ LLF +A+++QV+LV D L+ F + GLK + KS+ +
Sbjct: 125 LGTH*DF*KWPWYISHLFFPDNCLLFVKATSSQVKLVRDVLQQFCLT*GLKAGLQKSRFL 304
Query: 101 SSKGV 105
+ K V
Sbjct: 305 A*KNV 319
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.137 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,594,103
Number of Sequences: 63676
Number of extensions: 748723
Number of successful extensions: 4956
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 4829
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4936
length of query: 709
length of database: 12,639,632
effective HSP length: 104
effective length of query: 605
effective length of database: 6,017,328
effective search space: 3640483440
effective search space used: 3640483440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0108.16


