

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.10
(457 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
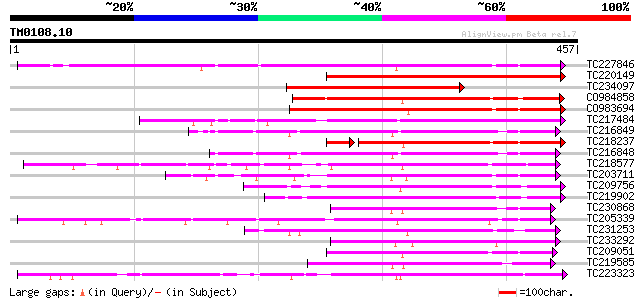
Score E
Sequences producing significant alignments: (bits) Value
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 306 1e-83
TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferas... 267 8e-72
TC234097 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin 5... 246 2e-65
CO984858 205 4e-53
CO983694 201 6e-52
TC217484 177 7e-45
TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransf... 174 1e-43
TC218237 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferas... 154 2e-41
TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 8... 160 9e-40
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 159 2e-39
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 152 2e-37
TC209756 152 4e-37
TC219902 similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase H... 149 3e-36
TC230868 weakly similar to UP|Q94AB5 (Q94AB5) AT3g46660/F12A12_1... 144 8e-35
TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucos... 144 1e-34
TC231253 weakly similar to UP|Q8S997 (Q8S997) Glucosyltransferas... 144 1e-34
TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, pa... 143 2e-34
TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferas... 141 7e-34
TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltr... 140 9e-34
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 137 1e-32
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 306 bits (785), Expect = 1e-83
Identities = 174/450 (38%), Positives = 268/450 (58%), Gaps = 8/450 (1%)
Frame = +2
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSDGFD 66
L++ YPAQGHINP +QFAKRL S G + T+ TT +T +N PN+T+ SDGFD
Sbjct: 2 LVLPYPAQGHINPLVQFAKRLASKGVK-ATVATTHYT----ANSINAPNITVEAISDGFD 166
Query: 67 DGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAEAARGLH 126
+ + L+ + G+ +++LI Q P TC++Y W + A+
Sbjct: 167 QAGFAQTNNNVQLFLASFRTNGSRTLSELIRKHQQTPSPSTCIVYDSFFPWVLDVAKQHG 346
Query: 127 LPTALLWVQPATVLDIIYYYFHGYHDC--LLSKIEFELPGLPFSLSPSDLPSFL-LPSND 183
+ A + A V +I HG+ + + +PGLP L LPSF+ P +
Sbjct: 347 IYGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLRVPGLP-PLDSRALPSFVRFPESY 523
Query: 184 SFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEE-FNVIPIGPLIPSAFLDGKD 242
++ QF L+ + VN+FEALE E ++ + E F IGP++PS +LDG+
Sbjct: 524 PAYMAMKLSQFSNLNNA--DWMFVNTFEALESEVLKGLTELFPAKMIGPMVPSGYLDGRI 697
Query: 243 LNDTSFGGDIFR-LSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPF 301
D +G +++ L+ C+ WL+ + +SVVY+SFGS V L++ QMEE+A L + G F
Sbjct: 698 KGDKGYGASLWKPLTEECSNWLESKPPQSVVYISFGSMVSLTEEQMEEVAWGLKESGVSF 877
Query: 302 LWVIREMEE---ELSCREELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSG 358
LWV+RE E L RE +++KG +VTWC+Q+E+L+H++ GCF+THCGWNST+ESL G
Sbjct: 878 LWVLRESEHGKLPLGYRESVKDKGLIVTWCNQLELLAHQATGCFVTHCGWNSTLESLSLG 1057
Query: 359 VPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEE 418
VP+V P+++DQ+ +AK ++++W++GV +E G+V E + L VM G++++E
Sbjct: 1058VPVVCLPQWADQLPDAKFLDEIWEVGVWPKE--DEKGIVRKQEFVQSLKDVM-EGQRSQE 1228
Query: 419 VRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
+RRNA K+K LA EA EGGSS+K++ F+
Sbjct: 1229IRRNANKWKKLAREAVGEGGSSDKHINQFV 1318
>TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferase like
protein, partial (27%)
Length = 820
Score = 267 bits (682), Expect = 8e-72
Identities = 133/195 (68%), Positives = 159/195 (81%), Gaps = 2/195 (1%)
Frame = +2
Query: 256 SNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEE-ELSC 314
SN EWLD Q E SVVYVSFG+ VL+ QM+E+ARALLD G+ FLWVIR+M+ E +C
Sbjct: 5 SNDYVEWLDSQPELSVVYVSFGTLAVLADRQMKELARALLDSGYLFLWVIRDMQGIEDNC 184
Query: 315 REELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNA 374
REELE++GK+V WCSQVEVLSH S+GCF+THCGWNSTMESL SGVPMVAFP+++DQ TNA
Sbjct: 185 REELEQRGKIVKWCSQVEVLSHGSLGCFVTHCGWNSTMESLGSGVPMVAFPQWTDQGTNA 364
Query: 375 KMVEDVWKIGVRMDHSVN-EDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEA 433
KMV+DVWK GVR+D VN E+G+VE EIRKCLD VMG+G K +E RRNA+K+K LA EA
Sbjct: 365 KMVQDVWKTGVRVDDKVNVEEGIVEAEEIRKCLDVVMGSGGKGQEFRRNADKWKCLAREA 544
Query: 434 GKEGGSSEKNLMAFL 448
EGGSS+ N+ FL
Sbjct: 545 VTEGGSSDSNMRTFL 589
>TC234097 similar to UP|Q9ZR25 (Q9ZR25) UDP-glucose:anthocyanin
5-O-glucosyltransferase, partial (30%)
Length = 435
Score = 246 bits (627), Expect = 2e-65
Identities = 119/144 (82%), Positives = 126/144 (86%), Gaps = 1/144 (0%)
Frame = +1
Query: 224 FNVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLS 283
FN+IPIGPLIPSAFLDGKD DTSFGGDIFR SN C EWLD + E SVVYVSFGSF VLS
Sbjct: 4 FNMIPIGPLIPSAFLDGKDPTDTSFGGDIFRPSNDCGEWLDSKPEMSVVYVSFGSFCVLS 183
Query: 284 KTQMEEIARALLDCGHPFLWVIREM-EEELSCREELEEKGKVVTWCSQVEVLSHRSVGCF 342
K QMEE+A ALLDCG PFLWV RE EEELSCREELE+KGK+V WCSQVEVLSHRSVGCF
Sbjct: 184 KKQMEELALALLDCGSPFLWVSREKEEEELSCREELEQKGKIVNWCSQVEVLSHRSVGCF 363
Query: 343 LTHCGWNSTMESLVSGVPMVAFPR 366
+THCGWNSTMESL SGVPM AFP+
Sbjct: 364 VTHCGWNSTMESLASGVPMFAFPQ 435
>CO984858
Length = 778
Score = 205 bits (521), Expect = 4e-53
Identities = 106/221 (47%), Positives = 150/221 (66%), Gaps = 2/221 (0%)
Frame = -2
Query: 229 IGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQME 288
IGP +PS FLD + +D +G F+ S C EWLD + + SVVYVSFGS +S+ QME
Sbjct: 774 IGPNVPSFFLDKQCEDDQDYGVTQFK-SEECVEWLDDKPKGSVVYVSFGSMATMSEEQME 598
Query: 289 EIARALLDCGHPFLWVIREMEEELSCR--EELEEKGKVVTWCSQVEVLSHRSVGCFLTHC 346
E+A L +C FLWV+R EE + E++ EKG VVTWCSQ++VL+H +VGCF+THC
Sbjct: 597 EVACCLRECSSYFLWVVRASEEIKLPKDFEKITEKGLVVTWCSQLKVLAHEAVGCFVTHC 418
Query: 347 GWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCL 406
GWNS +E+L GVP +A P +SDQ TNAK++ DVWKIG+R V+E +V ++ C+
Sbjct: 417 GWNSILETLCLGVPTIAIPCWSDQRTNAKLIADVWKIGIRT--PVDEKNIVRREALKHCI 244
Query: 407 DAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAF 447
+M ++ +E++ NA ++K LA+ A EGGSS +N++ F
Sbjct: 243 KEIM---DRDKEMKTNAIQWKTLAVRATAEGGSSYENIIEF 130
>CO983694
Length = 858
Score = 201 bits (511), Expect = 6e-52
Identities = 104/228 (45%), Positives = 146/228 (63%), Gaps = 5/228 (2%)
Frame = -1
Query: 226 VIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKT 285
++ IGP +P LD NDT + F++ + WL + SV+Y+SFGS V S
Sbjct: 858 ILMIGPTVPXXHLDKAVPNDTDNXXNXFQVDSSAISWLRQKPAGSVIYISFGSMVCFSSQ 679
Query: 286 QMEEIARALLDCGHPFLWVIREMEEELSCREELEE-----KGKVVTWCSQVEVLSHRSVG 340
QMEEIA L+ G FLWVI ++E + +E EE +G +V W Q+EVLS+ +VG
Sbjct: 678 QMEEIALGLMATGFNFLWVIPDLERKNLPKELGEEINACGRGLIVNWTPQLEVLSNHAVG 499
Query: 341 CFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGG 400
CF THCGWNST+E+L GVPMVA P+++DQ TNAK VEDVWK+G+R+ NE+G+V
Sbjct: 498 CFFTHCGWNSTLEALCLGVPMVALPQWTDQPTNAKFVEDVWKVGIRVKE--NENGIVTRE 325
Query: 401 EIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
E+ C+ VM + E+R NA+K+K LA+EA +GG+S+ N+ F+
Sbjct: 324 EVENCIRVVM-EKDLGREMRINAKKWKELAIEAVSQGGTSDNNINEFI 184
>TC217484
Length = 1651
Score = 177 bits (450), Expect = 7e-45
Identities = 120/355 (33%), Positives = 189/355 (52%), Gaps = 11/355 (3%)
Frame = +2
Query: 105 PFTCLIYTLLLHWAAEAARGLHLPTALLWVQPATVLDIIYYY--FHGYHDCLLSKIEFE- 161
P T ++ + L W A ++P A W A+ ++++ F + + K +
Sbjct: 197 PPTAILGCVELRWPIAVANRRNIPVAAFWTMSASFYSMLHHLDVFARHRGLTVDKDTMDG 376
Query: 162 ----LPGLPFSLSPSDLPSFLLPSNDSFIVSYFEEQFRELDVEANPTIL--VNSFEALEP 215
+PG+ S +DL + +L ND ++ E ++ AN +L V EA
Sbjct: 377 QAENIPGIS-SAHLADLRT-VLHENDQRVMQLALECISKVP-RANYLLLTTVQELEAETI 547
Query: 216 EAMRAVEEFNVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVS 275
E+++A+ F V PIGP IP L LN+ S+ +WLD Q +SV+Y+S
Sbjct: 548 ESLKAIFPFPVYPIGPAIPYLELGQNPLNNDH--------SHDYIKWLDSQPPESVLYIS 703
Query: 276 FGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEEELSCREELEEKGKVVTWCSQVEVLS 335
FGSF+ +S TQM++I AL +LWV R L +E+ +KG VV WC Q++VLS
Sbjct: 704 FGSFLSVSTTQMDQIVEALNSSEVRYLWVARANASFL--KEKCGDKGMVVPWCDQLKVLS 877
Query: 336 HRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHS-VNED 394
H SVG F +HCGWNST+E+L +GVPM+ FP F DQ+ N+ + D WK G +++ S ++ +
Sbjct: 878 HSSVGGFWSHCGWNSTLEALFAGVPMLTFPLFLDQVPNSSQIVDEWKNGSKVETSKLDSE 1057
Query: 395 GVVEGGEIRKCLDAVMG-NGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
+V +I + + M ++ +E+R A + K + + A GGSS NL AF+
Sbjct: 1058VIVAKEKIEELVKRFMDLQSQEGKEIRDRAREIKVMCLRAIAAGGSSYGNLDAFI 1222
>TC216849 weakly similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8,
partial (47%)
Length = 1125
Score = 174 bits (440), Expect = 1e-43
Identities = 107/312 (34%), Positives = 173/312 (55%), Gaps = 12/312 (3%)
Frame = +2
Query: 145 YYFHGYHDCLLSKIEFELPGLPFSLSPSDLPSFLLPSN-DSFIVSYFEEQFRELDVEANP 203
Y +GY D SK+++ +PG+ + D+P F+ ++ ++ ++ +F E + ++ N
Sbjct: 8 YLTNGYLD---SKVDW-IPGMK-NFRLKDIPDFIRTTDLNNVMLQFFIEVANK--IQRNT 166
Query: 204 TILVNSFEALEPEAMRAVEEF--NVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAE 261
TIL N+F+ LE + M A+ ++ PIGP P + TS G +++ C E
Sbjct: 167 TILFNTFDGLESDVMNALSSMFPSLYPIGPF-PLLLNQSPQSHLTSLGSNLWNEDLECLE 343
Query: 262 WLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIRE---------MEEEL 312
WL+ + +SVVYV+FGS V+S Q+ E A L + PFLW+IR + E
Sbjct: 344 WLESKESRSVVYVNFGSITVMSAEQLLEFAWGLANSKKPFLWIIRPDLVIGGSVILSSEF 523
Query: 313 SCREELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMT 372
E ++ + +WC Q +VL+H S+G FLTHCGWNST ES+ +GVPM+ +P F++Q T
Sbjct: 524 V--SETRDRSLIASWCPQEQVLNHPSIGVFLTHCGWNSTTESVCAGVPMLCWPFFAEQPT 697
Query: 373 NAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAME 432
N + + + W+IG+ +D S + E+ K ++ +M GEK +++R + K A E
Sbjct: 698 NCRYICNEWEIGMEIDTSAKRE------EVEKLVNELM-VGEKGKKMREKVMELKRKAEE 856
Query: 433 AGKEGGSSEKNL 444
K GG S NL
Sbjct: 857 VTKPGGCSYMNL 892
>TC218237 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (57%)
Length = 1176
Score = 154 bits (390), Expect(2) = 2e-41
Identities = 76/169 (44%), Positives = 116/169 (67%), Gaps = 2/169 (1%)
Frame = +3
Query: 282 LSKTQMEEIARALLDCGHPFLWVIREMEEELSCRE--ELEEKGKVVTWCSQVEVLSHRSV 339
L++ Q EE+A L D G F+WVIR+ ++ +E + EKG +V+WC Q++VL+H ++
Sbjct: 141 LNEEQTEELAWGLGDSGSYFMWVIRDCDKGKLPKEFADTSEKGLIVSWCPQLQVLTHEAL 320
Query: 340 GCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEG 399
GCFLTHCGWNST+E+L GVP++A P ++DQ+TNAK+++DVWKIGV+ +E +V
Sbjct: 321 GCFLTHCGWNSTLEALSLGVPVIAMPLWTDQITNAKLLKDVWKIGVKA--VADEKEIVRR 494
Query: 400 GEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFL 448
I C+ ++ EK E+++NA K+K LA EGG+S+KN+ F+
Sbjct: 495 ETITHCIKEIL-ETEKGNEIKKNAIKWKNLAKSYVDEGGNSDKNIAEFV 638
Score = 32.7 bits (73), Expect(2) = 2e-41
Identities = 14/23 (60%), Positives = 17/23 (73%)
Frame = +2
Query: 256 SNGCAEWLDLQAEKSVVYVSFGS 278
S C +WLD + + SVVYVSFGS
Sbjct: 62 SEACIKWLDEKPKGSVVYVSFGS 130
>TC216848 similar to UP|Q6VAB3 (Q6VAB3) UDP-glycosyltransferase 85A8, partial
(39%)
Length = 1113
Score = 160 bits (406), Expect = 9e-40
Identities = 100/296 (33%), Positives = 162/296 (53%), Gaps = 13/296 (4%)
Frame = +1
Query: 162 LPGLPFSLSPSDLPSFLLPSN-DSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRA 220
+PGL + DLP FL ++ + F++ +F E + V + + N+F LE +A+ A
Sbjct: 10 IPGLQ-NYRLKDLPDFLRTTDPNDFMLHFFIEVAEK--VPSASAVAFNTFHELERDAINA 180
Query: 221 VEEF--NVIPIGPLIPSAFLDGKDLNDT-SFGGDIFRLSNGCAEWLDLQAEKSVVYVSFG 277
+ ++ IGP +FLD S G ++++ GC +WL+ + +SVVYV+FG
Sbjct: 181 LPSMFPSLYSIGPF--PSFLDQSPHKQVPSLGSNLWKEDTGCLDWLESKEPRSVVYVNFG 354
Query: 278 SFVVLSKTQMEEIARALLDCGHPFLWVIRE---------MEEELSCREELEEKGKVVTWC 328
S V+S Q+ E A L + PFLW+IR + E E ++ + +WC
Sbjct: 355 SITVMSAEQLLEFAWGLANSKKPFLWIIRPDLVIGGSVILSSEFV--NETRDRSLIASWC 528
Query: 329 SQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMD 388
Q +VL+H S+G FLTHCGWNST ES+ +GVPM+ +P F+DQ TN + + + W+IG+ +D
Sbjct: 529 PQEQVLNHPSIGVFLTHCGWNSTTESICAGVPMLCWPFFADQPTNCRYICNEWEIGMEID 708
Query: 389 HSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNL 444
+ + E+ K ++ +M GEK +++ + + K A E + GG S NL
Sbjct: 709 TNAKRE------ELEKLVNELM-VGEKGKKMGQKTMELKKKAEEETRPGGGSYMNL 855
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 159 bits (402), Expect = 2e-39
Identities = 147/477 (30%), Positives = 223/477 (45%), Gaps = 44/477 (9%)
Frame = +1
Query: 12 PAQGHINPTLQFAKRLISSGAE-HVTLCTTLHTYRRISNK-------PTLPNLTLLPFSD 63
P H+ P L+F+KRL+ E H+T S+K PT+ ++ L P +
Sbjct: 25 PGFTHLVPILEFSKRLLHLHPEFHITCFIPSVGSSPTSSKAYVQTLPPTITSIFLPPIT- 201
Query: 64 GFDDGFKLTPDTDYSLYATELK---RRGTEFVTDLILSAAQEGKPFTCLIYTLLLHWAAE 120
L +D S+ A +++ ++ + + S K L+ + + A
Sbjct: 202 -------LDHVSDPSVLALQIELSVNLSLPYIREELKSLCSRAK-VVALVVDVFANGALN 357
Query: 121 AARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIE-----FELPGLPFSLSPSDLP 175
A+ L+L + + Q A +L + Y+Y + L S+ ++PG + DLP
Sbjct: 358 FAKELNLLSYIYLPQSAMLLSL-YFYSTKLDEILSSESRELQKPIDIPGC-VPIHNKDLP 531
Query: 176 SFLLPSNDSFIVSY--FEEQFRELDVEANPTILVNSFEALEPEAMRAVEEF-----NVIP 228
LP +D + Y F E+ + V + +N+F LE A+RA+EE + P
Sbjct: 532 ---LPFHDLSGLGYKGFLERSKRFHVPDG--VFMNTFLELESGAIRALEEHVKGKPKLYP 696
Query: 229 IGPLIPSAFLDGKDLNDTSFGGDIFRLSNG--CAEWLDLQAEKSVVYVSFGSFVVLSKTQ 286
+GP+I S G + NG C WLD Q SV+YVSFGS LS+ Q
Sbjct: 697 VGPIIQME----------SIGHE-----NGVECLTWLDKQEPNSVLYVSFGSGGTLSQEQ 831
Query: 287 MEEIARALLDCGHPFLWVIREMEEELSCR------------------EELEEKGKVV-TW 327
E+A L G FLWV+R +S E +++G VV +W
Sbjct: 832 FNELAFGLELSGKKFLWVVRAPSGVVSAGYLCAETKDPLEFLPHGFLERTKKQGLVVPSW 1011
Query: 328 CSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRM 387
Q++VL H + G FL+HCGWNS +ES+V GVP++ +P F++Q NA M+ D K+ +R
Sbjct: 1012APQIQVLGHSATGGFLSHCGWNSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKVALR- 1188
Query: 388 DHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNL 444
VNE G+VE EI K + +MG+ E E+R+ K A A KE GSS K L
Sbjct: 1189-PKVNESGLVEREEIAKVVRGLMGDKESL-EIRKRMGLLKIAAANAIKEDGSSTKTL 1353
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 152 bits (385), Expect = 2e-37
Identities = 113/348 (32%), Positives = 179/348 (50%), Gaps = 29/348 (8%)
Frame = +3
Query: 126 HLPTALLWVQPATVLDIIYYYFHGYHDCLLSK------IEFELPGLPFSLSPSDLPSFLL 179
++PT + A+ L ++ YY H L+ + ++ ++PGL +++ D P+
Sbjct: 99 NVPTYFYYTSGASTLALLLYY-PTIHPTLIEEKDTDQPLQIQIPGLS-TITADDFPNECK 272
Query: 180 PSNDSFIVSYFEEQFREL--DVEANPTILVNSFEALEPEAMRAVEEFNVIP-----IGPL 232
+SY + F ++ + I+VN+FEA+E EA+RA+ E +P +GP+
Sbjct: 273 DP-----LSYACQVFLQIAETMMGGAGIIVNTFEAIEEEAIRALSEDATVPPPLFCVGPV 437
Query: 233 IPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIAR 292
I + + G++ GC WL+LQ +SVV + FGS S+ Q++EIA
Sbjct: 438 ISAPY--GEE-------------DKGCLSWLNLQPSQSVVLLCFGSMGRFSRAQLKEIAI 572
Query: 293 ALLDCGHPFLWVIR-------EMEEELSCREEL--------EEKGKVVT-WCSQVEVLSH 336
L FLWV+R + EELS E L +EKG VV W Q +LSH
Sbjct: 573 GLEKSEQRFLWVVRTELGGADDSAEELSLDELLPEGFLERTKEKGMVVRDWAPQAAILSH 752
Query: 337 RSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGV 396
SVG F+THCGWNS +E++ GVPMVA+P +++Q N ++ K+ + + N+DG
Sbjct: 753 DSVGGFVTHCGWNSVLEAVCEGVPMVAWPLYAEQKMNRMVMVKEMKVALAVKE--NKDGF 926
Query: 397 VEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNL 444
V E+ + +M +K +E+R+ K K A EA EGG+S +L
Sbjct: 927 VSSTELGDRVRELM-ESDKGKEIRQRIFKMKMSAAEAMAEGGTSRASL 1067
>TC209756
Length = 1356
Score = 152 bits (383), Expect = 4e-37
Identities = 91/266 (34%), Positives = 146/266 (54%), Gaps = 6/266 (2%)
Frame = +1
Query: 189 YFEEQFRELD-VEANPTILVNSFEALEPEAMRAVEEFNVIPIGPLIPSAFLDGKDLNDTS 247
+F+ +E+ +E L N+ LEP A +F +PIGPL+ S D + ++
Sbjct: 127 FFDHLVQEMKTLELGEWWLCNTTYDLEPGAFSVSPKF--LPIGPLMES------DNSKSA 282
Query: 248 FGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIRE 307
F + C EWLD Q +SV+YVSFGS V+ Q +E+A AL PF+WV+R
Sbjct: 283 FWEE----DTTCLEWLDQQPPQSVIYVSFGSLAVMDPNQFKELALALDLLDKPFIWVVRP 450
Query: 308 MEEELS-----CREELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMV 362
+ + KGK+V W Q ++L+H ++ F++HCGWNST+E + +GVP +
Sbjct: 451 CNDNKENVNAYAHDFHGSKGKIVGWAPQKKILNHPALASFISHCGWNSTLEGICAGVPFL 630
Query: 363 AFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRN 422
+P +DQ + + DVWKIG+ +D +E+G++ EIRK +D ++ + E+++
Sbjct: 631 CWPCATDQYLDKSYICDVWKIGLGLDK--DENGIISREEIRKKVDQLLVD----EDIKAR 792
Query: 423 AEKFKGLAMEAGKEGGSSEKNLMAFL 448
+ K K + + EGG S KNL F+
Sbjct: 793 SLKLKDMTINNILEGGQSSKNLNFFM 870
>TC219902 similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (52%)
Length = 967
Score = 149 bits (375), Expect = 3e-36
Identities = 83/243 (34%), Positives = 132/243 (54%)
Frame = +1
Query: 206 LVNSFEALEPEAMRAVEEFNVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDL 265
L N+ LEP + ++ N++PIGPL+ S G + G + C WLD
Sbjct: 37 LCNTTNELEPGPLSSIP--NLVPIGPLLRSY---GDTIATAKSIGQYWEEDLSCMSWLDQ 201
Query: 266 QAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEEELSCREELEEKGKVV 325
Q SV+YV+FGSF + Q E+A + PFLWV+R+ + + E L KGK+V
Sbjct: 202 QPHGSVLYVAFGSFTHFDQNQFNELALGIDLTNRPFLWVVRQDNKRVYPNEFLGCKGKIV 381
Query: 326 TWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGV 385
+W Q +VLSH ++ CF+THCGWNST+E + +G+P++ +P F DQ+ N + D K+G+
Sbjct: 382 SWAPQQKVLSHPAIACFVTHCGWNSTIEGVSNGLPLLCWPYFGDQICNKTYICDELKVGL 561
Query: 386 RMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLM 445
D N G+V E+ + +D ++ + E ++ + + K M + G S +NL
Sbjct: 562 GFDSDKN--GLVSRMELERKVDQILND----ENIKSRSLELKDKVMNNIAKAGRSLENLN 723
Query: 446 AFL 448
F+
Sbjct: 724 RFV 732
>TC230868 weakly similar to UP|Q94AB5 (Q94AB5) AT3g46660/F12A12_180
(Glucosyltransferase-like protein), partial (38%)
Length = 765
Score = 144 bits (363), Expect = 8e-35
Identities = 81/191 (42%), Positives = 108/191 (56%), Gaps = 9/191 (4%)
Frame = +1
Query: 259 CAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIR-------EMEEE 311
C WLD Q SVVYVSFGS +S+ + EIA L + PFLWVIR E E
Sbjct: 7 CMSWLDQQDRNSVVYVSFGSIAAISEAEFLEIAWGLANSKQPFLWVIRPGLIHGSEWFEP 186
Query: 312 LSCR--EELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSD 369
L E L +G +V W Q +VLSH +VG F TH GWNST+ES+ GVPM+ P F+D
Sbjct: 187 LPSGFLENLGGRGYIVKWAPQEQVLSHPAVGAFWTHNGWNSTLESICEGVPMICMPCFAD 366
Query: 370 QMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGL 429
Q NAK VW++GV++ + ++ GE+ K + +M G++ E+R NA K
Sbjct: 367 QKVNAKYASSVWRVGVQLQNKLDR------GEVEKTIKTLM-VGDEGNEIRENALNLKEK 525
Query: 430 AMEAGKEGGSS 440
+ K+GGSS
Sbjct: 526 VNVSLKQGGSS 558
>TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose
glucosyltransferase, partial (55%)
Length = 1537
Score = 144 bits (362), Expect = 1e-34
Identities = 136/472 (28%), Positives = 218/472 (45%), Gaps = 38/472 (8%)
Frame = +2
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLH--TYRRISNKPTLPNLTLL----P 60
+L +P QGH+ P A+ G + T L+ T R K T ++ +L P
Sbjct: 38 MLFPFPGQGHLIPMSDMARAFNGRGVRTTIVTTPLNVATIRGTIGKETETDIEILTVKFP 217
Query: 61 FSD-GFDDGFKLT---PDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLLH 116
++ G +G + T P D L + R + L+L + +P CLI +
Sbjct: 218 SAEAGLPEGCENTESIPSPDLVLTFLKAIRMLEAPLEHLLL----QHRPH-CLIASAFFP 382
Query: 117 WAAEAARGLHLPTALLWVQPATVL---DIIYYYFHGYHDCLLSKIEFELPGLPFSLSPSD 173
WA+ +A L +P + L + + Y + + F +P LP + +
Sbjct: 383 WASHSATKLKIPRLVFHGTGVFALCASECVRLY-QPHKNVSSDTDPFIIPHLPGDIQMTR 559
Query: 174 L--PSFLLPSNDSFI-VSYFEEQFRELDVEANPTILVNSFEALEP-------EAMRAVEE 223
L P + D ++ ++ +E ++ A+ ++VNSF LE + + V+
Sbjct: 560 LLLPDYAKTDGDGETGLTRVLQEIKESEL-ASYGMIVNSFYELEQVYADYYDKQLLQVQG 736
Query: 224 FNVIPIGPLIPSAFLDGKDLNDTSFG-GDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVL 282
IGPL GK S GDI + WLD + SVVYV FGS
Sbjct: 737 RRAWYIGPLSLCNQDKGKRGKQASVDQGDILK-------WLDSKKANSVVYVCFGSIANF 895
Query: 283 SKTQMEEIARALLDCGHPFLWVIREMEEE--------LSCREELEEKGKVVT-WCSQVEV 333
S+TQ+ EIAR L D G F+WV+R +++ R E +G ++ W QV +
Sbjct: 896 SETQLREIARGLEDSGQQFIWVVRRSDKDDKGWLPEGFETRTTSEGRGVIIWGWAPQVLI 1075
Query: 334 LSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGV-----RMD 388
L H++VG F+THCGWNST+E++ +GVPM+ +P ++Q N K V D+ +IGV + +
Sbjct: 1076LDHQAVGAFVTHCGWNSTLEAVSAGVPMLTWPVSAEQFYNEKFVTDILQIGVPVGVKKWN 1255
Query: 389 HSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSS 440
V ++ + ++K L +M GE+AE +R A K +A A + GSS
Sbjct: 1256RIVGDN--ITSNALQKALHRIM-IGEEAEPMRNRAHKLAQMATTALQHNGSS 1402
>TC231253 weakly similar to UP|Q8S997 (Q8S997) Glucosyltransferase-12,
partial (31%)
Length = 841
Score = 144 bits (362), Expect = 1e-34
Identities = 97/264 (36%), Positives = 138/264 (51%), Gaps = 9/264 (3%)
Frame = +3
Query: 190 FEEQFRELDVEANPTILVNSFEALEPEAMRAVEEF--NVIPIGPL--IPSAFLDGKDLND 245
FEE A I++N+FE LEP + + V IGPL + + +
Sbjct: 15 FEETLAMTQASA---IILNTFEQLEPSIITKLATIFPKVYSIGPLHTLCKTMITTNSTSS 185
Query: 246 TSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVI 305
G + + C WLD Q KSV+YVSFG+ V LS Q+ E L++ PFLWVI
Sbjct: 186 PHKDGRLRKEDRSCITWLDHQKAKSVLYVSFGTVVNLSYEQLMEFWHGLVNSLKPFLWVI 365
Query: 306 -REMEEELSCREELE----EKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVP 360
+E+ + + ELE E+G +V W Q EVL++ +VG FLTHCGWNST+ES+ GVP
Sbjct: 366 QKELIIQKNVPIELEIGTKERGFLVNWAPQEEVLANPAVGGFLTHCGWNSTLESIAEGVP 545
Query: 361 MVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVR 420
M+ +P +DQ N++ V + WKIG+ M+ S D V +R ++ E++
Sbjct: 546 MLCWPSITDQTVNSRCVSEQWKIGLNMNGS--*DRFVVENMVRDIME--------NEDLM 695
Query: 421 RNAEKFKGLAMEAGKEGGSSEKNL 444
R+A A+ KE GSS NL
Sbjct: 696 RSANDVAKKALHGIKENGSSYHNL 767
>TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, partial (46%)
Length = 993
Score = 143 bits (360), Expect = 2e-34
Identities = 87/205 (42%), Positives = 119/205 (57%), Gaps = 19/205 (9%)
Frame = +1
Query: 259 CAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIREME-----EELS 313
C +WLDLQ KSVVYV FGS L +Q+ E+A AL D PF+WVIRE E+
Sbjct: 247 CLKWLDLQKSKSVVYVCFGSLCNLIPSQLVELALALEDTKRPFVWVIREGSKYQELEKWI 426
Query: 314 CREELEEKGK-----VVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFS 368
E EE+ K + W QV +LSH ++G FLTHCGWNST+E + +G+PM+ +P F+
Sbjct: 427 SEEGFEERTKGRGLIIRGWAPQVLILSHHAIGGFLTHCGWNSTLEGIGAGLPMITWPLFA 606
Query: 369 DQMTNAKMVEDVWKIGVRMDHSV-------NEDGV-VEGGEIRKCLDAVM-GNGEKAEEV 419
DQ N K+V V KIGV + V + GV V+ +I + + VM +GE+++E
Sbjct: 607 DQFLNEKLVTKVLKIGVSVGVEVPMKFGEEEKTGVLVKKEDINRAICMVMDDDGEESKER 786
Query: 420 RRNAEKFKGLAMEAGKEGGSSEKNL 444
R A K +A A + GGSS +L
Sbjct: 787 RERATKLSEMAKRAVENGGSSHLDL 861
>TC209051 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (35%)
Length = 1017
Score = 141 bits (355), Expect = 7e-34
Identities = 85/205 (41%), Positives = 113/205 (54%), Gaps = 19/205 (9%)
Frame = +2
Query: 256 SNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEEELSCR 315
S C WLD Q SV+YVSFGS LS+ Q+ E+A L G FLWV+R +S
Sbjct: 2 SGKCLSWLDKQPPCSVLYVSFGSGGTLSQNQINELASGLELSGQRFLWVLRAPSNSVSAA 181
Query: 316 ------------------EELEEKGKVV-TWCSQVEVLSHRSVGCFLTHCGWNSTMESLV 356
E +EKG VV +W QV+VL H SVG FL+HCGWNST+ES+
Sbjct: 182 YLEASKEDPLQFLPSGFLERTKEKGLVVASWAPQVQVLGHNSVGGFLSHCGWNSTLESVQ 361
Query: 357 SGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKA 416
GVP++ +P F++Q NA M+ D K+ +R NEDG+VE EI K + +M +GE+
Sbjct: 362 EGVPLITWPLFAEQRMNAVMLTDGLKVALR--PKFNEDGIVEKEEIAKVIKCLM-DGEEG 532
Query: 417 EEVRRNAEKFKGLAMEAGKEGGSSE 441
+R K A A K+G SS+
Sbjct: 533 IGMRERMGNLKDSAASALKDGSSSQ 607
>TC219585 weakly similar to UP|Q8L5C7 (Q8L5C7) UDP-glucuronosyltransferase ,
partial (50%)
Length = 935
Score = 140 bits (354), Expect = 9e-34
Identities = 76/207 (36%), Positives = 116/207 (55%), Gaps = 7/207 (3%)
Frame = +2
Query: 241 KDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHP 300
KD G ++++ + C +WLD SV+YV++GS V+S+ ++E A L + P
Sbjct: 32 KDKGFKVSGSNLWKNDSKCIQWLDQWEPSSVIYVNYGSITVMSEDHLKEFAWGLANSNLP 211
Query: 301 FLWVIRE---MEEELSCRE----ELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTME 353
FLW+ R M E + E++++G + WC Q +VLSH SVG FLTHCGWNST+E
Sbjct: 212 FLWIKRPDLVMGESTQLPQDFLDEVKDRGYITGWCPQEQVLSHPSVGVFLTHCGWNSTLE 391
Query: 354 SLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNG 413
+ GVPM+ +P F++Q TN + + W IG+ + V + V L M G
Sbjct: 392 GISGGVPMIGWPFFAEQQTNCRYICTTWGIGMDIKDDVKREEVT-------TLVKEMITG 550
Query: 414 EKAEEVRRNAEKFKGLAMEAGKEGGSS 440
E+ +E+R+ ++K A+EA GGSS
Sbjct: 551 ERGKEMRQKCLEWKKKAIEATDMGGSS 631
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 137 bits (345), Expect = 1e-32
Identities = 124/476 (26%), Positives = 214/476 (44%), Gaps = 33/476 (6%)
Frame = +1
Query: 7 LLIIYPAQGHINPTLQFAKRLISSG--AEHVTLCT-----TLHTYRRISN-------KPT 52
+LI +PAQGH+N L ++ ++S +V T TL + ISN P+
Sbjct: 49 VLIPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFHAFEVPS 228
Query: 53 LPNLTLLPFSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYT 112
+ P ++ D L P + S + E R+ L+ S + + K +I+
Sbjct: 229 FVSPPPNPNNEETDFPAHLLPSFEASSHLREPVRK-------LLHSLSSQAKR-VIVIHD 384
Query: 113 LLLHWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSKIEFELPGLPFSLSPS 172
++ A+ A + + T ++Y+ + + E+P + + +
Sbjct: 385 SVMASVAQDATNMPNVENYTFHSTCTFGTAVFYWDKMGRPLVDGMLVPEIPSMEGCFT-T 561
Query: 173 DLPSFLLPSNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEEFN----VIP 228
D +F++ D FR++ N + N+ A+E + +E F +
Sbjct: 562 DFMNFMIAQRD----------FRKV----NDGNIYNTSRAIEGAYIEWMERFTGGKKLWA 699
Query: 229 IGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQME 288
+GP P AF + KD + F C EWLD Q SV+YVSFG+ + Q++
Sbjct: 700 LGPFNPLAF-EKKDSKERHF----------CLEWLDKQDPNSVLYVSFGTTTTFKEEQIK 846
Query: 289 EIARALLDCGHPFLWVIREMEE------------ELS--CREELEEKGKVVT-WCSQVEV 333
+IA L F+WV+R+ ++ E S E +E G VV W Q+E+
Sbjct: 847 KIATGLEQSKQKFIWVLRDADKGDIFDGSEAKWNEFSNEFEERVEGMGLVVRDWAPQLEI 1026
Query: 334 LSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNE 393
LSH S G F++HCGWNS +ES+ GVP+ A+P SDQ N+ ++ +V KIG+ + +
Sbjct: 1027LSHTSTGGFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGLVVKNWAQR 1206
Query: 394 DGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFLA 449
+ +V + + +M E +++R A + K + + EGG S + +F+A
Sbjct: 1207NALVSASNVENAVRRLMETKE-GDDMRERAVRLKNVIHRSMDEGGVSRMEIDSFIA 1371
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,961,883
Number of Sequences: 63676
Number of extensions: 333673
Number of successful extensions: 1983
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 1852
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1875
length of query: 457
length of database: 12,639,632
effective HSP length: 100
effective length of query: 357
effective length of database: 6,272,032
effective search space: 2239115424
effective search space used: 2239115424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0108.10


