

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
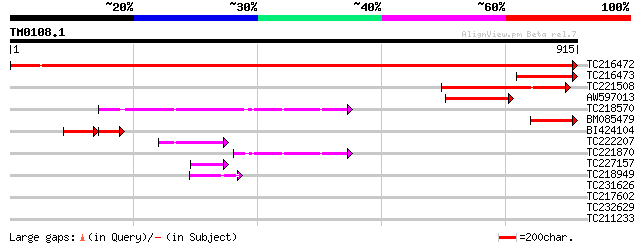
Query= TM0108.1
(915 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216472 UP|O63067 (O63067) Aspartokinase-homoserine dehydrogena... 1617 0.0
TC216473 homologue to UP|O63067 (O63067) Aspartokinase-homoserin... 184 2e-46
TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-... 173 3e-43
AW597013 similar to PIR|T06246|T06 aspartate kinase (EC 2.7.2.4)... 168 9e-42
TC218570 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctiona... 153 3e-37
BM085479 homologue to PIR|T06242|T062 aspartate kinase (EC 2.7.2... 140 3e-33
BI424104 homologue to PIR|T06246|T062 aspartate kinase (EC 2.7.2... 96 9e-33
TC222207 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctiona... 67 3e-11
TC221870 similar to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional ... 63 7e-10
TC227157 similar to UP|Q40329 (Q40329) Serine proteinase inhibit... 49 8e-06
TC218949 weakly similar to UP|Q7V6C2 (Q7V6C2) Uridylate kinase ... 44 4e-04
TC231626 similar to UP|Q9VXX2 (Q9VXX2) CG9101-PA, partial (10%) 33 0.74
TC217602 similar to UP|Q9SJW0 (Q9SJW0) Transfactor-like protein,... 32 1.7
TC232629 30 6.3
TC211233 weakly similar to GB|AAP21157.1|30102478|BT006349 At5g0... 29 8.2
>TC216472 UP|O63067 (O63067) Aspartokinase-homoserine dehydrogenase ,
complete
Length = 3148
Score = 1617 bits (4188), Expect = 0.0
Identities = 819/918 (89%), Positives = 868/918 (94%), Gaps = 3/918 (0%)
Frame = +2
Query: 1 MASLSASSLYHFSTISPSN---NTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGNQS 57
MAS SA+ + FS +SP++ ++ + Q QC PF S SHSLRK L+L PRG ++
Sbjct: 29 MASFSAA-VAQFSRVSPTHTPLHSHSHDTLFQSQCRPFFLSRTSHSLRKGLTL-PRGREA 202
Query: 58 PSTKISASLTDVSPSVLVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERK 117
PST + AS TDVSP+V +EE+QL KGETWSVHKFGGTC+G+SQRIKNV DI+L+DDSERK
Sbjct: 203 PSTTVRASFTDVSPNVSLEEKQLPKGETWSVHKFGGTCVGTSQRIKNVADIILKDDSERK 382
Query: 118 LVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLH 177
LVVVSAMSKVTDMMY+LI+KAQSRDESY ++L+AV EKHS TAHD+LDGD LATFLS+LH
Sbjct: 383 LVVVSAMSKVTDMMYDLIHKAQSRDESYTAALNAVSEKHSATAHDILDGDNLATFLSKLH 562
Query: 178 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLI 237
HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVI KNG DCKWMDTRDVLI
Sbjct: 563 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVITKNGTDCKWMDTRDVLI 742
Query: 238 VNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAI 297
VNPT SNQVDPDYLESE+RLE W+SLNPCKVIIATGFIASTPQ IPTTLKRDGSDFSAAI
Sbjct: 743 VNPTGSNQVDPDYLESEQRLEKWYSLNPCKVIIATGFIASTPQNIPTTLKRDGSDFSAAI 922
Query: 298 MGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 357
MGALF+ARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV
Sbjct: 923 MGALFKARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 1102
Query: 358 MQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMA 417
M+YGIPI+IRNIFNLSAPGTKICHPSVND+ED N++NFVKGFATIDNLAL+NVEGTGMA
Sbjct: 1103MRYGIPIMIRNIFNLSAPGTKICHPSVNDHEDSQNLQNFVKGFATIDNLALVNVEGTGMA 1282
Query: 418 GVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGR 477
GVPGTASAIFGAVKDVGANVIMISQASSEHS+CFAVPEKEVKAVAEALQSRFR ALDNGR
Sbjct: 1283GVPGTASAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGR 1462
Query: 478 LSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRE 537
LSQVA+IPNCSILAAVGQKMASTPGVSA+LFNALAKANINVRAIAQGCSEYNITVV+KRE
Sbjct: 1463LSQVAVIPNCSILAAVGQKMASTPGVSASLFNALAKANINVRAIAQGCSEYNITVVVKRE 1642
Query: 538 DCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILG 597
DC+KALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQAS LKEEFNIDLRVMGILG
Sbjct: 1643DCIKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASTLKEEFNIDLRVMGILG 1822
Query: 598 SKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHY 657
SKSMLLSD GIDLARWRELR+E GEVAN+EKFVQHVHGNHFIPNTA+VDCTADS IAG+Y
Sbjct: 1823SKSMLLSDVGIDLARWRELREERGEVANMEKFVQHVHGNHFIPNTALVDCTADSAIAGYY 2002
Query: 658 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 717
YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE
Sbjct: 2003YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 2182
Query: 718 TGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVI 777
TGD+ILQIEGIFSGTLSYIFNNFKDGRAFSEVV EAKEAGYTEPDPRDDLSGTDVARKVI
Sbjct: 2183TGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVSEAKEAGYTEPDPRDDLSGTDVARKVI 2362
Query: 778 ILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLR 837
ILARESGLKLELSNIPVESLVPEPL+ACASAQEFMQ LP+FDQ F KK E+AENAGEVLR
Sbjct: 2363ILARESGLKLELSNIPVESLVPEPLRACASAQEFMQELPKFDQEFTKKQEDAENAGEVLR 2542
Query: 838 YVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 897
YVGVVDVT+KKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA
Sbjct: 2543YVGVVDVTNKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 2722
Query: 898 GGIFSDILRLASYLGAPS 915
GGIFSDILRLASYLGAPS
Sbjct: 2723GGIFSDILRLASYLGAPS 2776
>TC216473 homologue to UP|O63067 (O63067) Aspartokinase-homoserine
dehydrogenase , partial (11%)
Length = 604
Score = 184 bits (467), Expect = 2e-46
Identities = 92/98 (93%), Positives = 95/98 (96%)
Frame = +2
Query: 818 FDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTR 877
FDQ F KK E+AENAGEVLRYVGVVDVT++KGVVELRRYKKDHPFAQLSGSDNIIAFTTR
Sbjct: 2 FDQEFTKKQEDAENAGEVLRYVGVVDVTNEKGVVELRRYKKDHPFAQLSGSDNIIAFTTR 181
Query: 878 RYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS 915
RYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS
Sbjct: 182 RYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS 295
>TC221508 similar to UP|Q8LF92 (Q8LF92) Homoserine dehydrogenase-like
protein, partial (60%)
Length = 912
Score = 173 bits (439), Expect = 3e-43
Identities = 90/210 (42%), Positives = 137/210 (64%), Gaps = 2/210 (0%)
Frame = +2
Query: 698 YEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAG 757
+E+TVGAGLP++++L ++ +GD + QI G SGTL Y+ + +DG+ S+VV++AK G
Sbjct: 50 HESTVGAGLPVIASLNRIISSGDPVHQIIGSLSGTLGYVMSEVEDGKPLSQVVRDAKSLG 229
Query: 758 YTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPL-QACASAQEFMQR-L 815
YTEPDPRDDL G DVARK +ILAR G ++ L +I +ESL P+ + + ++F+ R L
Sbjct: 230 YTEPDPRDDLGGMDVARKALILARILGHQINLDSIQIESLYPKEMGPGVMTVEDFLNRGL 409
Query: 816 PEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFT 875
D+ ++E+A + G VLRYV V + + V ++ K+ P +L GSDN++
Sbjct: 410 LLLDKDIQDRVEKAASNGNVLRYVCV--IMGSRCEVGIQELPKNSPLGRLRGSDNVLEIY 583
Query: 876 TRRYKDQPLIVRGPGAGAQVTAGGIFSDIL 905
TR Y QPL+++G GAG TA G+ +DI+
Sbjct: 584 TRCYSKQPLVIQGAGAGNDTTAAGVLADIV 673
>AW597013 similar to PIR|T06246|T06 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment), partial (12%)
Length = 334
Score = 168 bits (426), Expect = 9e-42
Identities = 86/110 (78%), Positives = 95/110 (86%)
Frame = +2
Query: 703 GAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPD 762
G GLPIVSTLRGLLETGD+ILQIEGIF GTLSYIFNNFKDGRA+SE E+KEA YT+P+
Sbjct: 5 GPGLPIVSTLRGLLETGDKILQIEGIFIGTLSYIFNNFKDGRAYSEESSESKEARYTDPN 184
Query: 763 PRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACASAQEFM 812
P DDLSGTD A K IILA+ESGLKLEL+NIP +SL PEPL ACA + EFM
Sbjct: 185 PIDDLSGTDAAIKAIILAKESGLKLELTNIPDDSLEPEPLLACA*SHEFM 334
>TC218570 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase, partial (75%)
Length = 1493
Score = 153 bits (387), Expect = 3e-37
Identities = 111/416 (26%), Positives = 206/416 (48%), Gaps = 6/416 (1%)
Frame = +1
Query: 144 SYVSSLDAVLEKHSLTAHDL-LDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTD 202
S + L + + H T L +DG +A L +L +L+ I + T+ D
Sbjct: 19 SSIEELCFIKDLHLRTVDQLGVDGSVIAKHLEELEQ-------LLKGIAMMKELTKRTQD 177
Query: 203 FVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNP---TSSNQVDPDYLESEKRLET 259
++V GE S ++ + + K G + D ++ + T+++ ++ Y KRL
Sbjct: 178 YLVSFGECMSTRIFAAYLNKIGVKARQYDAFEIGFITTDDFTNADILEATYPAVAKRLHG 357
Query: 260 WFSLNPCKVIIATGFIASTPQKIP-TTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYS 318
+ +P + I TGF+ + TTL R GSD +A +G ++ +W DVDGV +
Sbjct: 358 DWLSDPA-IAIVTGFLGKARKSCAVTTLGRGGSDLTATTIGKALGLPEIQVWKDVDGVLT 534
Query: 319 ADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTK 378
DP +A + L++ EA E++YFGA VLHP+++ P + IP+ ++N +N APGT
Sbjct: 535 CDPNIYPKAEPVPYLTFDEAAELAYFGAQVLHPQSMRPARESDIPVRVKNSYNPKAPGTL 714
Query: 379 ICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVI 438
I DM K + N+ ++++ T M G G + +F +++G +V
Sbjct: 715 I-----TKARDMS--KAVLTSIVLKRNVTMLDIASTRMLGQYGFLAKVFSIFEELGISVD 873
Query: 439 MISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN-GRLSQVAIIPNCSILAAVGQKM 497
++ A+SE S+ + ++ + E +Q H ++ +++ V ++ N SI++ +G
Sbjct: 874 VV--ATSEVSVSLTLDPSKLWS-RELIQQELDHVVEELEKIAVVNLLQNRSIISLIGNVQ 1044
Query: 498 ASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLS 553
S+ + F L + V+ I+QG S+ NI++V+ + + +RA+HS F+ S
Sbjct: 1045RSSL-ILEKAFRVLRTLGVTVQMISQGASKVNISLVVNDSEAEQCVRALHSAFFES 1209
>BM085479 homologue to PIR|T06242|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean, partial (8%)
Length = 347
Score = 140 bits (353), Expect = 3e-33
Identities = 68/75 (90%), Positives = 75/75 (99%)
Frame = +1
Query: 841 VVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGI 900
VVDV+++KGVVELRRYKKDHPFAQLSGSDNIIAFTTRRY++QPLIVRGPGAGAQVTAGGI
Sbjct: 1 VVDVSNEKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYRNQPLIVRGPGAGAQVTAGGI 180
Query: 901 FSDILRLASYLGAPS 915
F+DILRLA+YLGAPS
Sbjct: 181 FNDILRLATYLGAPS 225
>BI424104 homologue to PIR|T06246|T062 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment), partial (10%)
Length = 421
Score = 95.5 bits (236), Expect(2) = 9e-33
Identities = 46/56 (82%), Positives = 53/56 (94%)
Frame = +1
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDE 143
VHKFGGTC+G+SQRIKNV DI+L+DDS RKLVVVSAMSKV +MMY+LI+KAQSRDE
Sbjct: 1 VHKFGGTCVGTSQRIKNVADIILKDDSGRKLVVVSAMSKVKNMMYDLIHKAQSRDE 168
Score = 64.3 bits (155), Expect(2) = 9e-33
Identities = 28/41 (68%), Positives = 37/41 (89%)
Frame = +3
Query: 144 SYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLK 184
+Y+++LD+V EKH+ TAHD+LDG LA+FLS+LHHDISNLK
Sbjct: 168 AYIAALDSV*EKHNATAHDILDGHNLASFLSKLHHDISNLK 290
>TC222207 homologue to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase, partial (20%)
Length = 360
Score = 67.4 bits (163), Expect = 3e-11
Identities = 39/114 (34%), Positives = 64/114 (55%), Gaps = 1/114 (0%)
Frame = +3
Query: 241 TSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIP-TTLKRDGSDFSAAIMG 299
T+++ ++ Y KRL + + +P + I TGF+ + TTL R SD +A +G
Sbjct: 21 TNADILEATYPAVAKRLHSDWVSDPA-IPIVTGFLGKARKSCAVTTLGRG*SDLTATTIG 197
Query: 300 ALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRT 353
++ +W DVDGV + DP +A + L++ EA E++YFGA VLHP++
Sbjct: 198 KALGLPEIQVWKDVDGVLTCDPNICPQAKPVPYLTFDEAAELAYFGAXVLHPQS 359
>TC221870 similar to UP|Q9XHC5 (Q9XHC5) Precursor monofunctional
aspartokinase, partial (37%)
Length = 882
Score = 62.8 bits (151), Expect = 7e-10
Identities = 47/193 (24%), Positives = 96/193 (49%), Gaps = 1/193 (0%)
Frame = +2
Query: 362 IPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPG 421
IP+ ++N N APGT I DM K + N+ ++++ T M G G
Sbjct: 14 IPVRVKNXXNXKAPGTLIA-----KTRDMS--KALLTSIVLKRNVTMLDIVSTRMLGQFG 172
Query: 422 TASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN-GRLSQ 480
+ +F +++G +V ++ A+SE SI + ++ + E +Q + ++ +++
Sbjct: 173 FLAKVFSIFEELGISVDVV--ATSEVSISLTLDPSKLWS-RELIQQELDYVVEELEKIAV 343
Query: 481 VAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCV 540
V ++ SI++ +G S+ + F+ L + V+ I+QG S+ NI++V+ +
Sbjct: 344 VNLLKTRSIISLIGNVQRSSL-ILEKAFHVLRTLGVTVQMISQGASKVNISLVVNDSEAE 520
Query: 541 KALRAVHSRFYLS 553
+ +RA+H F+ S
Sbjct: 521 QCVRALHKAFFES 559
>TC227157 similar to UP|Q40329 (Q40329) Serine proteinase inhibitor, partial
(89%)
Length = 769
Score = 49.3 bits (116), Expect = 8e-06
Identities = 24/61 (39%), Positives = 36/61 (58%)
Frame = -3
Query: 293 FSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPR 352
+ ++G+ A V I TD+ GV +ADPR V AV L L++ EA E++ +G + H R
Sbjct: 185 YKPTLVGSSLGADYVLINTDISGVMTADPRIVDRAVALSHLNHHEALELAVYGTRMFHAR 6
Query: 353 T 353
T
Sbjct: 5 T 3
>TC218949 weakly similar to UP|Q7V6C2 (Q7V6C2) Uridylate kinase , partial
(92%)
Length = 1167
Score = 43.5 bits (101), Expect = 4e-04
Identities = 28/86 (32%), Positives = 46/86 (52%)
Frame = +1
Query: 291 SDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLH 350
+D +AA+ A A V T+VDGV+ DP++ +A + +TL+YQE +V+
Sbjct: 547 TDTAAALRCAEMNAEVVLKATNVDGVFDEDPKRNPQARLHETLTYQEVTSKD---LSVMD 717
Query: 351 PRTIIPVMQYGIPILIRNIFNLSAPG 376
I + IP++ +FNL+ PG
Sbjct: 718 MTAITLCQENNIPVI---VFNLNKPG 786
>TC231626 similar to UP|Q9VXX2 (Q9VXX2) CG9101-PA, partial (10%)
Length = 663
Score = 32.7 bits (73), Expect = 0.74
Identities = 20/80 (25%), Positives = 35/80 (43%)
Frame = +3
Query: 211 WSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNPCKVII 270
W + + V KNG + +W++ RD + N +++ D + +R W
Sbjct: 435 WFKECYATVTEKNGDEYEWVENRD*RLENAMRKIKIEKDIIIENRR--KWRERE------ 590
Query: 271 ATGFIASTPQKIPTTLKRDG 290
G S+ Q++ T L RDG
Sbjct: 591 CNGIA*SSTQQLETPLMRDG 650
>TC217602 similar to UP|Q9SJW0 (Q9SJW0) Transfactor-like protein, partial
(45%)
Length = 1744
Score = 31.6 bits (70), Expect = 1.7
Identities = 39/186 (20%), Positives = 78/186 (40%)
Frame = +2
Query: 697 FYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEA 756
F +A G P +T +G+L R++ + G+ + ++ + E + K+
Sbjct: 494 FVDAITQLGGPDRATPKGVL----RVMGVPGLTIYHVKSHLQKYRLAKYLPESPADGKDP 661
Query: 757 GYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLP 816
+ D +SG D + + I L++++ V+ + E L+ ++ R+
Sbjct: 662 KDEKRMSGDSISGADSSSGMPI---NDALRMQME---VQKRLHEQLEV---QKQLQMRIE 814
Query: 817 EFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTT 876
+ K +EE + G L TS+ + L K++HP ++ SGS + +A T
Sbjct: 815 AQGKYLQKIIEEQQKLGSTL-------TTSE--TLPLSHDKQNHPQSEASGSSDALASTV 967
Query: 877 RRYKDQ 882
K Q
Sbjct: 968 SPLKKQ 985
>TC232629
Length = 514
Score = 29.6 bits (65), Expect = 6.3
Identities = 16/46 (34%), Positives = 23/46 (49%)
Frame = +1
Query: 803 QACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKK 848
Q +AQ ++ + FA KL+ E LRYVG VD++ K
Sbjct: 142 QTWITAQNILREKLITEDCFAWKLQAGSKEEEALRYVGGVDISFSK 279
>TC211233 weakly similar to GB|AAP21157.1|30102478|BT006349
At5g02970/F9G14_280 {Arabidopsis thaliana;} , partial
(7%)
Length = 497
Score = 29.3 bits (64), Expect = 8.2
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +2
Query: 78 QQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLED 112
Q L +++ V KFGG C S+R++ + D + ED
Sbjct: 362 QNLINEDSYVVRKFGGPCSKVSKRLRFLNDFLPED 466
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,098,733
Number of Sequences: 63676
Number of extensions: 442888
Number of successful extensions: 2143
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 2123
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2141
length of query: 915
length of database: 12,639,632
effective HSP length: 106
effective length of query: 809
effective length of database: 5,889,976
effective search space: 4764990584
effective search space used: 4764990584
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0108.1


