

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.8
(1558 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
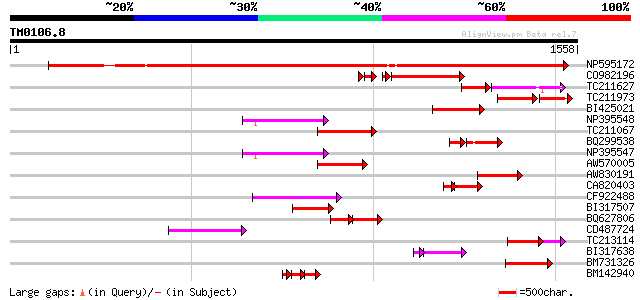
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 1149 0.0
CO982196 244 1e-72
TC211627 113 5e-48
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 116 8e-45
BI425021 160 4e-39
NP395548 reverse transcriptase [Glycine max] 160 4e-39
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 149 9e-36
BQ299538 105 5e-35
NP395547 reverse transcriptase [Glycine max] 145 1e-34
AW570005 144 3e-34
AW830191 142 9e-34
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 109 2e-33
CF922488 137 4e-32
BI317507 128 2e-29
BQ627806 76 2e-27
CD487724 119 1e-26
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 115 1e-25
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 94 2e-22
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 102 2e-21
BM142940 59 5e-21
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 1149 bits (2973), Expect = 0.0
Identities = 621/1440 (43%), Positives = 867/1440 (60%), Gaps = 10/1440 (0%)
Frame = +1
Query: 107 RVDIPMFNGNDAYGWVTKVERFFRLSRVEEAEKIEMVMIAMEDRALGWFQWWEEQTLERA 166
++D P F+G + W+ K E+FF +A+++ + + ++ + W+Q ++ +
Sbjct: 295 KLDFPRFDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWYQMLQKTEPFSS 474
Query: 167 WEPFKQALFRRFQPALLQNPFGPLLSVKQKGSVMEYKENFELLAAPMRNADREVLKGVFL 226
W+ F +AL F P+ P L + Q +V EY F L + E + F+
Sbjct: 475 WQAFTRALELDFGPSAYDCPRATLFKLNQSATVNEYYMQFTALVNRVDGLSAEAILDCFV 654
Query: 227 NGLQEEIKAEMKLYPADDLAELMDRALLLEEKNTAMRGGKPKEEDKRGWKEKGGVGGRYY 286
+GLQEEI ++K L + + A L EEK T+ PK
Sbjct: 655 SGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTS----PPK------------------ 768
Query: 287 SSTGDSMGRIANSYVNFQSKGGTGNQDNEGKSLQNKGGTGNQDTEGKQPEKKWNGGQRLT 346
+ T ++ R S + K NQ N+ T K + ++++
Sbjct: 769 TKTFSNLARNFTSNTSATQKYPPTNQKNDNPKPNLPPLLPTPST--KPFNLRNQNIKKIS 942
Query: 347 QTELQERSRKGLCFKCGDKWGKEHICSMKNYQLILMEVEEDEEEEEIFEEAEDGEFVLEG 406
E+Q R K LC+ C +K+ H C N Q++L+++EE +E++ + E ++
Sbjct: 943 PAEIQLRREKNLCYFCDEKFSPAHKCP--NRQVMLLQLEETDEDQTDEQVMVTEEANMDD 1116
Query: 407 KVLQLSLNSKEGLTSNRSFKVKGKIGNREVLILIDCGATINFISQDLVVELEIPVIATSE 466
LSLN+ G + + G++G V IL+D G++ NFI + L++PV
Sbjct: 1117 DTHHLSLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAPN 1296
Query: 467 YVVEVGNGAKERNSGVCKNLKLEVQGIPIMQHFFILGLGGTEVVLGMDWLASLGNIEANF 526
V VGNG G+ + L L +QG + ++L + G +V+LG WLA+LG A++
Sbjct: 1297 LRVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADY 1476
Query: 527 QELIIQWVSQGQKMVLQGEPSVCKVAANWKSIK-ITEQQEAEGYYLSYEYQKE-EEKTEA 584
L +++ + + LQGE + A + + + E + QKE E T
Sbjct: 1477 AALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKEVPEDTLK 1656
Query: 585 EVPEGMRK----ILEEYPEVFQEPKGLPPRRTTDHAIQLQEGASIPNIRPYRYPFYQKNE 640
++P + +L Y +VF P LPP+R DHAI L++G+ +RPYRYP QK++
Sbjct: 1657 DLPTNIDPELAILLHTYAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQ 1836
Query: 641 IEKLVKEMLNSGIIRHSTSPFSSPAILVKKKDGGWRFCVDYRAINKATIPDKFPIPIIDE 700
IEK+++EML GII+ S SPFS P +LVKKKDG WRFC DYRA+N T+ D FP+P +DE
Sbjct: 1837 IEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDE 2016
Query: 701 LLDEIGAAVVFSKLDLKSGYHQIRMKEEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQA 760
LLDE+ A FSKLDL+SGYHQI ++ ED KTAFRTH GHYE+LV+PFGLTNAP+TFQ
Sbjct: 2017 LLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQC 2196
Query: 761 LMNQVLRPYLRKFVLVFFYDILIYSKNEELHKDHLRIVLQVLKENNLVANQKKCSFGQPE 820
LMN++ + LRKFVLVFF DILIYS + + H HL VLQ LK++ L A KCSFG E
Sbjct: 2197 LMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTE 2376
Query: 821 IIYLGHVISQAGVAADPSKIKDMLDWPIPKEVKGLRGFLGLTGYYRRFVKNYSKLAQPLN 880
+ YLGH +S GV+ + +K++ +LDWP P VK LRGFLGLTGYYRRF+K+Y+ +A PL
Sbjct: 2377 VDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAGPLT 2556
Query: 881 QLLKKNSFQWTEEATQAFVKLKEVMTTVPVLVPPNFDKPFILETDASGKGLGAVLMQEGR 940
LL+K+SF W EA AFVKLK+ MT PVL P+F +PFILETDASG G+GAVL Q G
Sbjct: 2557 DLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQNGH 2736
Query: 941 PVAYMSKTLSDRAQAKSVYERELMAVVLAVQKWRHYLLGSQFVIHTDQRSLRFLADQRIM 1000
P+AY SK L+ R Q +S Y REL+A+ A+ K+RHYLLG++F+I TDQRSL+ L DQ +
Sbjct: 2737 PIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQ 2916
Query: 1001 GEEQQKWMSKLMGYDFEIKYKPGIENKAADALSRKLQFSAISSVQCAEWADLEAEILGDE 1060
EQQ W+ K +GYDF+I+YKPG +N+AADALSR A S +L A ++ D
Sbjct: 2917 TPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSRMFML-AWSEPHSIFLEELRARLISDP 3093
Query: 1061 RYRKVLQELATQGNSAIGYQLKRGRLLYKDRIVLPKGSTKILTVLKEFHDTALGGHAGIF 1120
+ K L E QG A Y ++ G L +KDR+V+P + +L+E+H + +GGHAGI
Sbjct: 3094 -HLKQLMETYKQGADASHYTVREGLLYWKDRVVIPAEEEIVNKILQEYHSSPIGGHAGIT 3270
Query: 1121 RTYKRISALFYWEGMKLDIQNYVQKCEVCQRNKYEALNPAGFLQPLPIPSQGWTDISMDF 1180
RT R+ A FYW M+ D++ Y+QKC +CQ+ K PAG LQPLPIP Q W D++MDF
Sbjct: 3271 RTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIPQQVWEDVAMDF 3450
Query: 1181 IGGLPKAMGKDTILVVVDRFTKYAHFIALSHPYNAKEIAEVFIKEVVKLHGFPTSIVSDR 1240
I GLP + G I+VV+DR TKYAHFI L YN+K +AE F+ +VKLHG P SIVSDR
Sbjct: 3451 ITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRSIVSDR 3630
Query: 1241 DRVFLSTFWSEMFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGSKPKQWPKWL 1300
DRVF STFW +FKL GT L SSAYHPQ+DGQ+EV+N+C+E YLRC T PK W K L
Sbjct: 3631 DRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHPKGWVKAL 3810
Query: 1301 SWAEFWYNTNYHSAIKTTPFKALYGRESPVIFKGNDSLTSVDEVEKWTAERNLILEELKS 1360
WAEFWYNT YH ++ TPF+ALYGRE P + + S+ EV + +R+ +L +LK
Sbjct: 3811 PWAEFWYNTAYHMSLGMTPFRALYGREPPTLTRQACSIDDPAEVREQLTDRDALLAKLKI 3990
Query: 1361 NLEKAQNRMRQQANKHRRDVQYEVGDLVYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIA 1420
NL +AQ M++QA+K R DV +++GD V +K+QPY+ S R NQKLS RY+GP+ ++A
Sbjct: 3991 NLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKVLA 4170
Query: 1421 KINPAAYKLQLPEGSQVHPVFHISLLKKAVNAGVQSQPLPAALT-EEWELKVEPEAIMDT 1479
KI AYKL+LP +++HPVFH+S L K N Q LP LT E ++P I+ +
Sbjct: 4171 KIGDVAYKLELPSAARIHPVFHVSQL-KPFNGTAQDPYLPLPLTVTEMGPVMQPVKILAS 4347
Query: 1480 R---ENRDGDLEVLIRWKDLPTFEDSWEDFSKLLDQFPNHQLEDKLSLQGGRDVANPSSR 1536
R + ++L++W++ E +WED + +P LEDK+ +G +V N SR
Sbjct: 4348 RIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKASYPTFNLEDKVVFKGEGNVTNGMSR 4527
>CO982196
Length = 812
Score = 244 bits (623), Expect(4) = 1e-72
Identities = 118/201 (58%), Positives = 149/201 (73%)
Frame = +1
Query: 1048 EWADLEAEILGDERYRKVLQELATQGNSAIGYQLKRGRLLYKDRIVLPKGSTKILTVLKE 1107
E AD E EI ++ Q + T+ GY ++ G+L +KDR+VL K STKI +LKE
Sbjct: 208 ELADWEEEIQAYLELYEIYQGILTKTTKKPGYAIRGGKLYFKDRLVLSKNSTKIPLLLKE 387
Query: 1108 FHDTALGGHAGIFRTYKRISALFYWEGMKLDIQNYVQKCEVCQRNKYEALNPAGFLQPLP 1167
D+ LGGH+G FRT+KR++ + +W+GMK ++YV CE+C+RNK L+PAG L LP
Sbjct: 388 LQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACEICRRNKTSTLSPAGLL*LLP 567
Query: 1168 IPSQGWTDISMDFIGGLPKAMGKDTILVVVDRFTKYAHFIALSHPYNAKEIAEVFIKEVV 1227
IP++ WTDISMDFIGGLPKA GKD ILVVVDR TKYAHF ALSHPY AKE+AE+FIKE+V
Sbjct: 568 IPTKVWTDISMDFIGGLPKAQGKDNILVVVDRLTKYAHFFALSHPYTAKEVAELFIKELV 747
Query: 1228 KLHGFPTSIVSDRDRVFLSTF 1248
+LHGFP SIVSD R+F+S F
Sbjct: 748 RLHGFPASIVSDXXRLFMSLF 810
Score = 44.7 bits (104), Expect(4) = 1e-72
Identities = 19/34 (55%), Positives = 26/34 (75%)
Frame = +2
Query: 974 RHYLLGSQFVIHTDQRSLRFLADQRIMGEEQQKW 1007
RHY +G +F+I T+ RS +FL +QR+M EEQ KW
Sbjct: 53 RHYPVGKKFIIRTN*RSSKFLNEQRLMSEEQFKW 154
Score = 25.0 bits (53), Expect(4) = 1e-72
Identities = 11/22 (50%), Positives = 15/22 (68%)
Frame = +3
Query: 1024 IENKAADALSRKLQFSAISSVQ 1045
+ N ++ LSR+ FSAIS VQ
Sbjct: 135 VRNSSSGLLSRQFSFSAISMVQ 200
Score = 21.9 bits (45), Expect(4) = 1e-72
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +3
Query: 958 VYERELMAVVLAV 970
+YERELM VVL V
Sbjct: 6 MYERELMDVVLPV 44
>TC211627
Length = 1034
Score = 113 bits (283), Expect(2) = 5e-48
Identities = 71/215 (33%), Positives = 118/215 (54%), Gaps = 10/215 (4%)
Frame = +3
Query: 1323 LYGRESPVIFKGNDSLTSVDEVEKWTAERNLILEELKSNLEKAQNRMRQQANKHRRDVQY 1382
+YG+ P + + ++V+ V+ I L L+K Q+ M++ A+ HRRD+ +
Sbjct: 243 MYGKPPPALPLYSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTF 422
Query: 1383 EVGDLVYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIAKINPAAYKLQLPEGSQVHPVFH 1442
+GD VY+++ PY+ S+ + + KLS R+YGPY I A++ AY+LQLP S++HP+FH
Sbjct: 423 NIGDWVYVRL*PYRQTSI-QSTYTKLSKRFYGPYQIQARVGQVAYRLQLPPTSKIHPIFH 599
Query: 1443 ISLLKKAVNAGVQSQPLPAAL-------TEEWELKVEPEAIMDTRENRDGD---LEVLIR 1492
+SLLK V P+P L T L V+P +D + + +VL++
Sbjct: 600 VSLLK------VHHGPIPPELLALPPFSTTNHPL-VQPLQFLDWKMDESTTPPIPQVLVQ 758
Query: 1493 WKDLPTFEDSWEDFSKLLDQFPNHQLEDKLSLQGG 1527
W +L + +WE +++L D + LEDK+ Q G
Sbjct: 759 WTNLAPEDTTWESWTQLKDIY---DLEDKVCFQTG 854
Score = 98.2 bits (243), Expect(2) = 5e-48
Identities = 44/79 (55%), Positives = 58/79 (72%)
Frame = +2
Query: 1243 VFLSTFWSEMFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGSKPKQWPKWLSW 1302
+F+S W E+F ++GTKL+FS+AYHPQTDGQTEV+NR +E YLR P+ W K+LS
Sbjct: 2 IFISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSL 181
Query: 1303 AEFWYNTNYHSAIKTTPFK 1321
AE YNT+ HS I +PF+
Sbjct: 182 AE*CYNTSVHSGIGFSPFE 238
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 116 bits (290), Expect(2) = 8e-45
Identities = 56/109 (51%), Positives = 77/109 (70%)
Frame = +1
Query: 1341 VDEVEKWTAERNLILEELKSNLEKAQNRMRQQANKHRRDVQYEVGDLVYLKIQPYKLKSL 1400
++EV K R+ +L L+ NL K+ + M NK RRD++Y VGD V+LK+QPY+ +SL
Sbjct: 13 LEEVNKLIIARDGLLATLRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYRRRSL 192
Query: 1401 AKRSNQKLSPRYYGPYPIIAKINPAAYKLQLPEGSQVHPVFHISLLKKA 1449
AKR N+KLSPR+Y P+ + K+ AYKL LP ++HPVFH+SLLKKA
Sbjct: 193 AKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPSHIKIHPVFHVSLLKKA 339
Score = 84.7 bits (208), Expect(2) = 8e-45
Identities = 40/92 (43%), Positives = 63/92 (68%)
Frame = +3
Query: 1455 QSQPLPAALTEEWELKVEPEAIMDTRENRDGDLEVLIRWKDLPTFEDSWEDFSKLLDQFP 1514
QSQPLP L+E+W+L+ ++++D+RE + G+++VLI+WK+LP E+SWE +KL + F
Sbjct: 339 QSQPLPPMLSEDWKLQTYSDSVLDSRELQPGNVKVLIQWKNLPPSENSWESVAKLQEIFS 518
Query: 1515 NHQLEDKLSLQGGRDVANPSSRPRFGNVYARR 1546
+ LEDK+SL GG + +P VY R+
Sbjct: 519 IYHLEDKVSLLGG-GIDKHKHKPPIPKVYTRK 611
>BI425021
Length = 426
Score = 160 bits (405), Expect = 4e-39
Identities = 75/142 (52%), Positives = 99/142 (68%)
Frame = -1
Query: 1163 LQPLPIPSQGWTDISMDFIGGLPKAMGKDTILVVVDRFTKYAHFIALSHPYNAKEIAEVF 1222
L PLP+P + W D+SMDFI GLP G TI VVV+RF+K H L + A +A +F
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLF 247
Query: 1223 IKEVVKLHGFPTSIVSDRDRVFLSTFWSEMFKLAGTKLKFSSAYHPQTDGQTEVVNRCVE 1282
+ V+KLHGFP SIVSDRD +F+S FW ++F+L+GT L+ SSAYHPQTDGQTEV+NR +E
Sbjct: 246 LNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIE 67
Query: 1283 TYLRCVTGSKPKQWPKWLSWAE 1304
YLR +P+ +++ W E
Sbjct: 66 QYLRAFVHGRPRNLGRFIPWVE 1
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 160 bits (405), Expect = 4e-39
Identities = 93/254 (36%), Positives = 136/254 (52%), Gaps = 19/254 (7%)
Frame = +1
Query: 641 IEKLVKEMLNSGIIRH-STSPFSSPAILVKKKDG------------------GWRFCVDY 681
+ K V ++L G+I S S + SP ++V KK+G W+ C+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 682 RAINKATIPDKFPIPIIDELLDEIGAAVVFSKLDLKSGYHQIRMKEEDIPKTAFRTHEGH 741
R +N+AT D FP+P +D++L+ + + LD GY+QI + +D K AF G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 742 YEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIYSKNEELHKDHLRIVLQV 801
+ Y +PFGL NAP+TFQ M + + K + VF D ++ + E L +VLQ
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 802 LKENNLVANQKKCSFGQPEIIYLGHVISQAGVAADPSKIKDMLDWPIPKEVKGLRGFLGL 861
E NLV N +KC F E I LGH IS G+ D +KI + P P VKG+R FLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 862 TGYYRRFVKNYSKL 875
+YRRF+K+++K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 149 bits (376), Expect = 9e-36
Identities = 74/163 (45%), Positives = 104/163 (63%), Gaps = 1/163 (0%)
Frame = +1
Query: 847 PIPKEVKGLRGFLGLTGYYRRFVKNYSKLAQPLNQLLKKN-SFQWTEEATQAFVKLKEVM 905
P K V +R F GL +YRRFV N+S +A PLN+L+KKN +F W E+ QAF LKE +
Sbjct: 97 PTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKL 276
Query: 906 TTVPVLVPPNFDKPFILETDASGKGLGAVLMQEGRPVAYMSKTLSDRAQAKSVYERELMA 965
T PVL P+F K F LE DASG G+ AVL+Q G P+AY S+ L Y++EL A
Sbjct: 277 TKAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYA 456
Query: 966 VVLAVQKWRHYLLGSQFVIHTDQRSLRFLADQRIMGEEQQKWM 1008
++ A Q W H+L+ +FVIH+D +SL+++ + + + KW+
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWV 585
Score = 33.1 bits (74), Expect = 0.98
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = +2
Query: 821 IIYLGHVISQAGVAADPSKIKDMLDWPIPKEVKGLRG 857
I + G V+ + GV DP KIK + +WP P +V + G
Sbjct: 17 IFFSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEILG 127
>BQ299538
Length = 426
Score = 105 bits (261), Expect(2) = 5e-35
Identities = 48/99 (48%), Positives = 67/99 (67%)
Frame = +3
Query: 1254 KLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGSKPKQWPKWLSWAEFWYNTNYHS 1313
KL GT LK S++YHP DGQT VN C+ET+LRC +PK +WLSWAE+WYNTN+H+
Sbjct: 135 KLPGTYLKMSTSYHP*IDGQT--VNHCLETFLRCFVADQPKM*VQWLSWAEYWYNTNFHA 308
Query: 1314 AIKTTPFKALYGRESPVIFKGNDSLTSVDEVEKWTAERN 1352
+ TTPF+ +YGR+ PV+ + V+ V + +R+
Sbjct: 309 STGTTPFEVVYGRKPPVLNRFLPGEVRVEAVRRELQDRD 425
Score = 63.2 bits (152), Expect(2) = 5e-35
Identities = 25/44 (56%), Positives = 35/44 (78%)
Frame = +1
Query: 1209 LSHPYNAKEIAEVFIKEVVKLHGFPTSIVSDRDRVFLSTFWSEM 1252
L HPY+A+ +AE+F KEVV LHG P S++SD D +F+S+FW E+
Sbjct: 1 LKHPYSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 145 bits (367), Expect = 1e-34
Identities = 89/254 (35%), Positives = 133/254 (52%), Gaps = 19/254 (7%)
Frame = +1
Query: 641 IEKLVKEMLNSGIIRH-STSPFSSPAILVKKKDG------------------GWRFCVDY 681
+ K V ++L +G+I S S + SP +V KK G WR C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 682 RAINKATIPDKFPIPIIDELLDEIGAAVVFSKLDLKSGYHQIRMKEEDIPKTAFRTHEGH 741
R +N+AT D +P+P +D++L + + LD SGY+QI + +D KTAF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 742 YEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIYSKNEELHKDHLRIVLQV 801
+ Y +PFGL NA +TFQ M + + K + VF D + + +L VLQ
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 802 LKENNLVANQKKCSFGQPEIIYLGHVISQAGVAADPSKIKDMLDWPIPKEVKGLRGFLGL 861
+++NLV N +KC F E I LGH IS+ G+ K+ + P P VKG+ FLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 862 TGYYRRFVKNYSKL 875
G+YRRF+K+++K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>AW570005
Length = 413
Score = 144 bits (363), Expect = 3e-34
Identities = 71/137 (51%), Positives = 89/137 (64%)
Frame = -2
Query: 847 PIPKEVKGLRGFLGLTGYYRRFVKNYSKLAQPLNQLLKKNSFQWTEEATQAFVKLKEVMT 906
P P+ + LRGFL LTG+YRRF+K Y+ +A PL+ LL K+SF W+ EA AF LK V+T
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDSFVWSPEADVAFQALKNVVT 233
Query: 907 TVPVLVPPNFDKPFILETDASGKGLGAVLMQEGRPVAYMSKTLSDRAQAKSVYERELMAV 966
VL P+F KPF +ETDASG +GAVL QEG P+A+ SK + S Y EL A+
Sbjct: 232 NTLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAI 53
Query: 967 VLAVQKWRHYLLGSQFV 983
V+KWR YLLG V
Sbjct: 52 TNVVKKWRQYLLGHHLV 2
>AW830191
Length = 372
Score = 142 bits (359), Expect = 9e-34
Identities = 65/123 (52%), Positives = 94/123 (75%)
Frame = +3
Query: 1285 LRCVTGSKPKQWPKWLSWAEFWYNTNYHSAIKTTPFKALYGRESPVIFKGNDSLTSVDEV 1344
LRC+TG+KP+QWPK LSWAEFW+NTNY++++K TPFK LYG + P + KG ++ +EV
Sbjct: 6 LRCLTGTKPQQWPKRLSWAEFWFNTNYNNSLKLTPFKVLYGCDPPHLLKGAIISSTAEEV 185
Query: 1345 EKWTAERNLILEELKSNLEKAQNRMRQQANKHRRDVQYEVGDLVYLKIQPYKLKSLAKRS 1404
T +R+ +L +LK NL KAQN+M+ A++ RR + VGD VYLK+QPY+ +SLA+++
Sbjct: 186 NVMTNDRDQMLHDLKGNLAKAQNQMK-YADRSRRSIPLNVGDWVYLKLQPYRQRSLARKT 362
Query: 1405 NQK 1407
N+K
Sbjct: 363 NEK 371
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 109 bits (272), Expect(2) = 2e-33
Identities = 53/77 (68%), Positives = 62/77 (79%), Gaps = 1/77 (1%)
Frame = -3
Query: 1222 FIKEVVKLHGFPTSIVSDRDRVFL-STFWSEMFKLAGTKLKFSSAYHPQTDGQTEVVNRC 1280
FIKE VKLHG +SIVSD DR+FL S FW+E+FK+ GTKLKFS AYHPQ D T+VVNRC
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 1281 VETYLRCVTGSKPKQWP 1297
+E L+C+T SK KQWP
Sbjct: 155 IEMNLQCLTTSKRKQWP 105
Score = 53.1 bits (126), Expect(2) = 2e-33
Identities = 22/34 (64%), Positives = 30/34 (87%)
Frame = -1
Query: 1193 ILVVVDRFTKYAHFIALSHPYNAKEIAEVFIKEV 1226
I+V+V RFTKYAHF+ LSHPY AKE++EV +K++
Sbjct: 421 IMVIVYRFTKYAHFVVLSHPYLAKEVSEVLLKKL 320
>CF922488
Length = 741
Score = 137 bits (345), Expect = 4e-32
Identities = 87/246 (35%), Positives = 132/246 (53%), Gaps = 1/246 (0%)
Frame = +3
Query: 668 VKKKDGGWRFCVDYRAINKATIPDKFPIPIIDELLDEIGAAVVFSKLDLKSGYHQIRMKE 727
V K+DG CVDYR +N A+ DKFP+P I+ L+D + FS +D SGY+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 728 EDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIYSKN 787
ED+ KT F T G + Y + FGL N +T+Q M + + K + V+ D+++ S+
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 788 EELHKDHLRIVLQVLKENNLVANQKKCSFGQPEIIYLGHVISQAGVAADPSKIKDMLDWP 847
EE H +LR + + L++ L N KC F L + S G+ D +K+K +L+
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 848 IPKEVKGLRGFLGLTGYYRRFVKNYSKLAQPLNQLLKKNSF-QWTEEATQAFVKLKEVMT 906
P K ++GFLG Y RF+ +PL LL KN F +W + AF ++K+ +
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 907 TVPVLV 912
VLV
Sbjct: 723 NPHVLV 740
>BI317507
Length = 359
Score = 128 bits (321), Expect = 2e-29
Identities = 63/114 (55%), Positives = 82/114 (71%), Gaps = 1/114 (0%)
Frame = -1
Query: 778 FYDILIYSKNEELHKDHLRIVLQVLKENNLVANQKKCSFGQPEIIYLGHVISQAGVAADP 837
FY+ILIYS + + H HL VL VLK+ LVAN+KKC F Q I YLGHVIS+ VA D
Sbjct: 359 FYNILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDS 180
Query: 838 SKIKDMLDWPIPKEVKGLRGFLGLTGYYRRFVKNYSKLA-QPLNQLLKKNSFQW 890
+K+K +++WP+PK VK + FL LTGYYR+F+K+Y KLA +PL L K + F+W
Sbjct: 179 NKVKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKW 18
>BQ627806
Length = 435
Score = 76.3 bits (186), Expect(2) = 2e-27
Identities = 36/83 (43%), Positives = 52/83 (62%)
Frame = +1
Query: 941 PVAYMSKTLSDRAQAKSVYERELMAVVLAVQKWRHYLLGSQFVIHTDQRSLRFLADQRIM 1000
P+ + + S Y REL A+ +AV+KWR YLLG FVI TD RSL+ L Q +
Sbjct: 181 PLPFSFMPFCSKLLRASTYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQ 360
Query: 1001 GEEQQKWMSKLMGYDFEIKYKPG 1023
EQQ ++++LMG+D+ I+Y+ G
Sbjct: 361 TPEQQIYLARLMGFDYTIQYRAG 429
Score = 66.2 bits (160), Expect(2) = 2e-27
Identities = 31/63 (49%), Positives = 43/63 (68%)
Frame = +3
Query: 882 LLKKNSFQWTEEATQAFVKLKEVMTTVPVLVPPNFDKPFILETDASGKGLGAVLMQEGRP 941
LL K+ F W EEA +AF +LK + PVL P+F+ F++ETDASG G+GA+L Q P
Sbjct: 3 LLVKDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHP 182
Query: 942 VAY 944
+A+
Sbjct: 183 LAF 191
>CD487724
Length = 676
Score = 119 bits (298), Expect = 1e-26
Identities = 67/216 (31%), Positives = 117/216 (54%), Gaps = 2/216 (0%)
Frame = +3
Query: 436 VLILIDCGATINFISQDLVVELEIPVIATSEYVVEVGNGAKERNSGVCKNLKLEVQGIPI 495
++ L+D G+T NF+ Q LV +L +P +T V VGNG + + +C+ + + +Q I
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTTICEAIPISIQNIEF 203
Query: 496 MQHFFILGLGGTEVVLGMDWLASLGNIEANFQELIIQWVSQGQKMVLQGEPSVCKVAANW 555
+ H ++L + G +VLG+ WL +LG I ++ L +Q+ Q + + L+GE N
Sbjct: 204 LVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGLLNH 383
Query: 556 KSIKITEQ--QEAEGYYLSYEYQKEEEKTEAEVPEGMRKILEEYPEVFQEPKGLPPRRTT 613
++ Q + ++++ + + +P+ ++ +L+++ +FQ P+GLPP R T
Sbjct: 384 HQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFSALFQ*PQGLPPARET 563
Query: 614 DHAIQLQEGASIPNIRPYRYPFYQKNEIEKLVKEML 649
DH I L + N+R Y YP Y NEIE V ML
Sbjct: 564 DHHIHLLP*SEPVNMRLY*YPHY*NNEIEHQVNLML 671
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 115 bits (288), Expect = 1e-25
Identities = 56/98 (57%), Positives = 71/98 (72%)
Frame = +3
Query: 1369 MRQQANKHRRDVQYEVGDLVYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIAKINPAAYK 1428
M+ A++ RR V VGD VYLK+QPY+LKSLAK+ N+KLSPR+YGPY I +I A++
Sbjct: 3 MKAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFE 182
Query: 1429 LQLPEGSQVHPVFHISLLKKAVNAGVQSQPLPAALTEE 1466
L LP ++HPVFH SLLKKAV A QPLP L+E+
Sbjct: 183 LDLPPARKIHPVFHASLLKKAVAATANPQPLPLMLSED 296
Score = 59.3 bits (142), Expect = 1e-08
Identities = 29/72 (40%), Positives = 43/72 (59%)
Frame = +2
Query: 1456 SQPLPAALTEEWELKVEPEAIMDTRENRDGDLEVLIRWKDLPTFEDSWEDFSKLLDQFPN 1515
S P+ + +EL+V P + N +G EVLI+ +DLP FE +WE + +QFP+
Sbjct: 263 SSTTPSDVI*RFELRVFPAEVKAVHNNSNGIAEVLIQLEDLPDFEATWESVEVIKEQFPS 442
Query: 1516 HQLEDKLSLQGG 1527
LEDK++L GG
Sbjct: 443 FHLEDKVTLLGG 478
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 94.0 bits (232), Expect(2) = 2e-22
Identities = 50/117 (42%), Positives = 64/117 (53%)
Frame = -2
Query: 1137 LDIQNYVQKCEVCQRNKYEALNPAGFLQPLPIPSQGWTDISMDFIGGLPKAMGKDTILVV 1196
L+ + C CQ KYE L PL +P + W D+S+DFI GL ILVV
Sbjct: 353 LECAQMLPNCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPYHVHTAILVV 174
Query: 1197 VDRFTKYAHFIALSHPYNAKEIAEVFIKEVVKLHGFPTSIVSDRDRVFLSTFWSEMF 1253
VD F+K H L + A +A +FI V KLHG P S+VSD D +F+S FW E+F
Sbjct: 173 VDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQELF 3
Score = 32.0 bits (71), Expect(2) = 2e-22
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = -1
Query: 1111 TALGGHAGIFRTYKRISALFYWEGMKLDI 1139
T GGH GI +T +S YW GM+ D+
Sbjct: 420 TTTGGHTGIAKTLA*LSKNIYWFGMRTDV 334
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 102 bits (253), Expect = 2e-21
Identities = 53/133 (39%), Positives = 84/133 (62%), Gaps = 2/133 (1%)
Frame = +1
Query: 1362 LEKAQNRMRQQANKHRRDVQYEVGDLVYLKIQPYKLKSLAKRSNQKLSPRYYGPYPIIAK 1421
LE+AQ+ M + AN HRR VGD VYLKI+P++ S+ R + KL+ R+YGPY ++ +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 1422 INPAAYKLQLPEGSQVHPVFHISLLKKAVNAGVQSQPLPAALTEEWEL--KVEPEAIMDT 1479
+ A++LQLP +++HPVFH+S LK+A+ + LP L + EL V+ I +
Sbjct: 205 VGAVAFQLQLPSEARIHPVFHVSQLKRALGNHQAQEELPPDLEHQAELYFPVQILKIREV 384
Query: 1480 RENRDGDLEVLIR 1492
++ + + +VLIR
Sbjct: 385 QKQHEVERQVLIR 423
>BM142940
Length = 422
Score = 58.9 bits (141), Expect(3) = 5e-21
Identities = 26/45 (57%), Positives = 35/45 (77%)
Frame = -3
Query: 809 ANQKKCSFGQPEIIYLGHVISQAGVAADPSKIKDMLDWPIPKEVK 853
AN+KKCSFG + YL H+IS GV+ADP K++ M+ WPIPKE++
Sbjct: 156 ANRKKCSFGCKKFEYLVHMISVKGVSADPKKLEVMVAWPIPKEMR 22
Score = 43.9 bits (102), Expect(3) = 5e-21
Identities = 22/39 (56%), Positives = 25/39 (63%)
Frame = -1
Query: 774 VLVFFYDILIYSKNEELHKDHLRIVLQVLKENNLVANQK 812
+ F YDILIYS NE H+ HL+IVLQ KE LV K
Sbjct: 260 ICFFSYDILIYSINEGKHEKHLKIVLQTFKEAKLVPTGK 144
Score = 38.1 bits (87), Expect(3) = 5e-21
Identities = 17/25 (68%), Positives = 21/25 (84%)
Frame = -2
Query: 749 FGLTNAPSTFQALMNQVLRPYLRKF 773
F LTNA STFQALMN++L+P+L F
Sbjct: 421 FSLTNALSTFQALMNELLKPFLCNF 347
Score = 35.0 bits (79), Expect = 0.26
Identities = 19/33 (57%), Positives = 24/33 (72%)
Frame = -2
Query: 752 TNAPSTFQALMNQVLRPYLRKFVLVFFYDILIY 784
TN STFQALM ++L+P+L KF VFF I +Y
Sbjct: 322 TNVLSTFQALMTELLQPFLCKF--VFFPMISLY 230
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,696,880
Number of Sequences: 63676
Number of extensions: 869508
Number of successful extensions: 6713
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 6022
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6486
length of query: 1558
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1448
effective length of database: 5,635,272
effective search space: 8159873856
effective search space used: 8159873856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0106.8


