

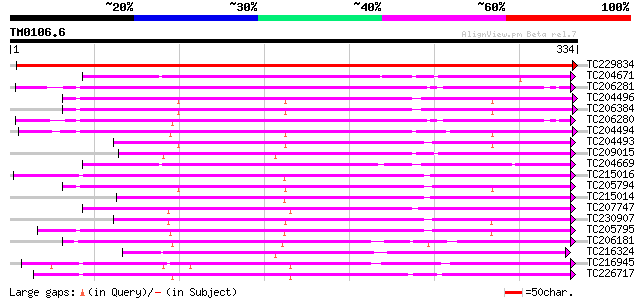
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.6
(334 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229834 similar to UP|PE26_ARATH (O22862) Probable peroxidase 2... 488 e-138
TC204671 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2... 213 1e-55
TC206281 UP|Q9ZRG5 (Q9ZRG5) Peroxidase (Fragment) , complete 211 5e-55
TC204496 homologue to UP|O23961 (O23961) Peroxidase precursor, p... 209 2e-54
TC206384 UP|O23961 (O23961) Peroxidase precursor, complete 208 3e-54
TC206280 UP|Q9ZRG6 (Q9ZRG6) Peroxidase (Fragment) , complete 205 2e-53
TC204494 similar to UP|O23961 (O23961) Peroxidase precursor, par... 205 3e-53
TC204493 UP|O22443 (O22443) Seed coat peroxidase precursor , co... 204 6e-53
TC209015 similar to UP|PE72_ARATH (Q9FJZ9) Peroxidase 72 precurs... 204 6e-53
TC204669 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2... 203 8e-53
TC215016 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (... 203 1e-52
TC205794 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), ... 203 1e-52
TC215014 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (... 198 2e-51
TC207747 weakly similar to UP|Q9M4Z5 (Q9M4Z5) Peroxidase prx12 p... 197 4e-51
TC230907 similar to UP|O24081 (O24081) Peroxidase1A precursor ,... 196 2e-50
TC205795 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), ... 194 6e-50
TC206181 similar to UP|Q7XYR7 (Q7XYR7) Class III peroxidase, par... 192 2e-49
TC216324 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, complete 190 7e-49
TC216945 similar to UP|Q43854 (Q43854) Peroxidase precursor , p... 189 1e-48
TC226717 homologue to UP|Q9ZTW8 (Q9ZTW8) Peroxidase, partial (85%) 189 2e-48
>TC229834 similar to UP|PE26_ARATH (O22862) Probable peroxidase 26 precursor
(Atperox P26) (ATP50) , partial (84%)
Length = 1150
Score = 488 bits (1257), Expect = e-138
Identities = 234/330 (70%), Positives = 268/330 (80%)
Frame = +3
Query: 5 MCVAFLLVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKS 64
MCV F LV +A A P L WHYYK N C DAE YVR+QV LFWK D+S
Sbjct: 30 MCVGFPLVALVVVSMCYGLADAEVKTPNLRWHYYKVTNRCHDAEEYVRHQVNLFWKNDRS 209
Query: 65 ITAKLLRLVYSDCFITGCDASILLDEGPNTEKKAPQNRGLGAFVLIDNIKTFVERQCPGV 124
ITAKLLRLVY+DCF+TGCDASILLDEG N EKKA QNRGLG F +ID IK +E +CPG
Sbjct: 210 ITAKLLRLVYADCFVTGCDASILLDEGANPEKKAAQNRGLGGFAVIDKIKAVLESRCPGT 389
Query: 125 VSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASVDIPSPSVSWQEALAYFKSRG 184
VSCADIL LATRDAV+LAGG GYPV TGRKDGM+SDAASVD+PSPSVS Q+ L YFKSR
Sbjct: 390 VSCADILHLATRDAVKLAGGAGYPVLTGRKDGMKSDAASVDLPSPSVSLQKVLEYFKSRN 569
Query: 185 LNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQS 244
LN LDM TLLGAHT+GRTHCS+I DRLYNYNG+G DPSM+ L+++RKLCPPRKKGQ+
Sbjct: 570 LNELDMTTLLGAHTMGRTHCSFIVDRLYNYNGSGKPDPSMSVTSLESLRKLCPPRKKGQA 749
Query: 245 DPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKKS 304
DPLV+LNP+SGSSY FTESYY+R+L+HE VLG+DQQLL DDTK+I+EEFA G +DF+KS
Sbjct: 750 DPLVHLNPESGSSYNFTESYYRRVLSHETVLGVDQQLLYSDDTKQISEEFAVGFEDFRKS 929
Query: 305 FAVSMYNMGNIKVLTGNQGQIRQNCRFINK 334
FA SMY MGN +VLTGNQG+IR+ CR+ NK
Sbjct: 930 FATSMYKMGNYRVLTGNQGEIRRYCRYTNK 1019
>TC204671 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (90%)
Length = 1185
Score = 213 bits (542), Expect = 1e-55
Identities = 125/296 (42%), Positives = 171/296 (57%), Gaps = 6/296 (2%)
Frame = +2
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPNTEKKAPQNRG 103
C AE V++ V D ++ A LLR+ + DCF+ GCDAS+L+ G TE+ A N G
Sbjct: 176 CPRAESIVKSTVTTHVNSDSTLAAGLLRMHFHDCFVQGCDASVLI-AGSGTERTAFANLG 352
Query: 104 LGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAAS 163
L F +ID+ K +E CPGVVSCADIL LA RD+V L+GG Y V TGR+DG S A+
Sbjct: 353 LRGFEVIDDAKKQLEAACPGVVSCADILALAARDSVVLSGGLSYQVLTGRRDGRISQASD 532
Query: 164 V-DIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDP 222
V ++P+P S F ++GLN D+ TL+GAHTIG T C + ++RLYN+ G DP
Sbjct: 533 VSNLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGAHTIGTTACQFFSNRLYNFTANG-PDP 709
Query: 223 SMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLL 282
S++ +FL ++ LCP + G V L D+GS KF SYY + N +L DQ L
Sbjct: 710 SIDPSFLSQLQSLCP--QNGDGSKRVAL--DTGSQTKFDLSYYSNLRNSRGILQSDQALW 877
Query: 283 NGDDTKEITEEFAAGLQ-----DFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
+ TK + + ++ F F SM MGNI++ TG G+IR+ C IN
Sbjct: 878 SDASTKTTVQRYLGLIRGLLGLTFNVEFGKSMVKMGNIELKTGTDGEIRKICSAIN 1045
>TC206281 UP|Q9ZRG5 (Q9ZRG5) Peroxidase (Fragment) , complete
Length = 1129
Score = 211 bits (536), Expect = 5e-55
Identities = 131/334 (39%), Positives = 190/334 (56%), Gaps = 4/334 (1%)
Frame = +3
Query: 4 SMCVAFL-LVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFD 62
++ VAFL L++ F+ +S+S L +YY C D E V VK D
Sbjct: 42 AVMVAFLNLIIMFSVVSTSK---------SLSLNYYS--KTCPDVECIVAKAVKDATARD 188
Query: 63 KSITAKLLRLVYSDCFITGCDASILLD-EGPN-TEKKAPQNRGLGAFVLIDNIKTFVERQ 120
K++ A LLR+ + DCF+ GCDAS+LL+ +G N EK P N L AF +ID K +E
Sbjct: 189 KTVPAALLRMHFHDCFVRGCDASVLLNSKGSNKAEKDGPPNVSLHAFYVIDAAKKALEAS 368
Query: 121 CPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASV-DIPSPSVSWQEALAY 179
CPGVVSCADIL LA RDAV L+GGP + V GRKDG S A+ +P+P+ + +
Sbjct: 369 CPGVVSCADILALAARDAVFLSGGPTWDVPKGRKDGRTSKASETRQLPAPTFNLSQLRQS 548
Query: 180 FKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPR 239
F RGL+ D+ L G HT+G +HCS +R++N+N T DPS+N +F + +CP +
Sbjct: 549 FSQRGLSGEDLVALSGGHTLGFSHCSSFKNRIHNFNATHDVDPSLNPSFATKLISICPLK 728
Query: 240 KKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQ 299
+ ++ ++P S+ F +YY+ IL + + DQ LL+ DTK + +FA +
Sbjct: 729 NQAKNAG-TSMDP---STTTFDNTYYRLILQQKGLFSSDQVLLDNPDTKNLVAKFATSKK 896
Query: 300 DFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
F +FA SM M +I G Q ++R++CR IN
Sbjct: 897 AFYDAFAKSMIKMSSI---NGGQ-EVRKDCRVIN 986
>TC204496 homologue to UP|O23961 (O23961) Peroxidase precursor, partial (97%)
Length = 2193
Score = 209 bits (531), Expect = 2e-54
Identities = 118/310 (38%), Positives = 178/310 (57%), Gaps = 7/310 (2%)
Frame = +3
Query: 32 KLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEG 91
+L+ +Y+ + C VR V+ K D + A L+RL + DCF+ GCDAS+LL+
Sbjct: 945 QLDPSFYR--DTCPKVHSIVREVVRNVSKKDPRMLASLIRLHFHDCFVQGCDASVLLNNT 1118
Query: 92 PNTEKKA---PQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYP 148
E + P N L ++++IKT VE+ CPGVVSCADIL LA+ + L GGP +
Sbjct: 1119 ATIESEQQALPNNNSLRGLDVVNDIKTAVEQACPGVVSCADILTLASEISSILGGGPDWK 1298
Query: 149 VFTGRKDGMRSDA--ASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSY 206
V GR+D + ++ A+ ++P+P + + A F +GL+ D+ L GAHT GR HCS+
Sbjct: 1299 VPLGRRDSLTANRTLANQNLPAPFFNLTQLKAAFAVQGLDTTDLVALSGAHTFGRAHCSF 1478
Query: 207 ITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYK 266
I RLYN++GTG DP+++ +L +R++CP P +N D + K Y+
Sbjct: 1479 ILGRLYNFSGTGKPDPTLDTTYLQQLRQICP-----NGGPNNLVNFDPVTPDKIDRVYFS 1643
Query: 267 RILNHEAVLGIDQQLLN--GDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQ 324
+ + +L DQ+L + G DT I F++ F +F SM MGNI VLTGN+G+
Sbjct: 1644 NLQVKKGLLQSDQELFSTPGADTIPIVNRFSSDQNVFFDAFEASMIKMGNIGVLTGNKGE 1823
Query: 325 IRQNCRFINK 334
IR++C F+NK
Sbjct: 1824 IRKHCNFVNK 1853
>TC206384 UP|O23961 (O23961) Peroxidase precursor, complete
Length = 1251
Score = 208 bits (529), Expect = 3e-54
Identities = 117/310 (37%), Positives = 178/310 (56%), Gaps = 7/310 (2%)
Frame = +1
Query: 32 KLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEG 91
+L+ +Y+ + C VR V+ K D + A L+RL + DCF+ GCDAS+LL+
Sbjct: 106 QLDPSFYR--DTCPRVHSIVREVVRNVSKKDPRMLASLIRLHFHDCFVQGCDASVLLNNT 279
Query: 92 PNTEKKA---PQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYP 148
E + P N L ++++IKT VE+ CPGVVSCADIL LA+ + L GGP +
Sbjct: 280 ATIESEQQALPNNNSLRGLDVVNDIKTAVEKACPGVVSCADILTLASEISSVLGGGPHWK 459
Query: 149 VFTGRKDGMRSDA--ASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSY 206
V GR+D + ++ A+ ++P+P + A F +GL+ D+ L GAHT GR HC++
Sbjct: 460 VPLGRRDSLTANRNLANQNLPAPFFNLSRLKAAFAVQGLDTTDLVALSGAHTFGRAHCNF 639
Query: 207 ITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYK 266
I DRLYN++GTG DP+++ +L +R++CP P +N D + K Y+
Sbjct: 640 ILDRLYNFSGTGKPDPTLDTTYLQQLRQICP-----NGGPNNLVNFDPVTPDKIDRVYFS 804
Query: 267 RILNHEAVLGIDQQLLN--GDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQ 324
+ + +L DQ+L + G DT I F++ + F +F SM MGNI VLTG +G+
Sbjct: 805 NLQVKKGLLQSDQELFSTPGADTIPIVNRFSSDQKVFFDAFEASMIKMGNIGVLTGKKGE 984
Query: 325 IRQNCRFINK 334
IR++C F+NK
Sbjct: 985 IRKHCNFVNK 1014
>TC206280 UP|Q9ZRG6 (Q9ZRG6) Peroxidase (Fragment) , complete
Length = 1174
Score = 205 bits (522), Expect = 2e-53
Identities = 125/333 (37%), Positives = 188/333 (55%), Gaps = 3/333 (0%)
Frame = +3
Query: 4 SMCVAFLLVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDK 63
++ VAFL ++ F+ +S++ + L +YY C + E V VK DK
Sbjct: 63 AVMVAFLNLIIFSVVSTTGKS--------LSLNYYA--KTCPNVEFIVAKAVKDATARDK 212
Query: 64 SITAKLLRLVYSDCFITGCDASILLDEGPNT--EKKAPQNRGLGAFVLIDNIKTFVERQC 121
++ A +LR+ + DCF+ GCDAS+LL+ N EK P N L AF +I K +E C
Sbjct: 213 TVPAAILRMHFHDCFVRGCDASVLLNSKGNNKAEKDGPPNVSLHAFYVIVAAKKALEASC 392
Query: 122 PGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASV-DIPSPSVSWQEALAYF 180
PGVVSCADIL LA RDAV L+GGP + V GRKDG S A+ +P+P+ + + F
Sbjct: 393 PGVVSCADILALAARDAVFLSGGPTWDVPKGRKDGRTSKASETRQLPAPTFNLSQLRQSF 572
Query: 181 KSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRK 240
RGL+ D+ L G HT+G +HCS +R++N+N T DPS+N +F + +CP +
Sbjct: 573 SQRGLSGEDLVALSGGHTLGFSHCSSFKNRIHNFNATHDVDPSLNPSFAAKLISICPLKN 752
Query: 241 KGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQD 300
+ ++ ++P S+ F +YY+ IL + + DQ LL+ DTK + +FA +
Sbjct: 753 QAKNAG-TSMDP---STTTFDNTYYRLILQQKGLFSSDQVLLDNPDTKNLVTKFATSKKA 920
Query: 301 FKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
F ++FA SM M +I G Q ++R++CR IN
Sbjct: 921 FYEAFAKSMIRMSSI---NGGQ-EVRKDCRMIN 1007
>TC204494 similar to UP|O23961 (O23961) Peroxidase precursor, partial (93%)
Length = 1326
Score = 205 bits (521), Expect = 3e-53
Identities = 123/336 (36%), Positives = 190/336 (55%), Gaps = 7/336 (2%)
Frame = +3
Query: 6 CVAFLLVVAFAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSI 65
C F+++ A L P ++ KLE +YK C V V+ + D +
Sbjct: 84 CFGFIVIGLVAVLGGLPFSS----NAKLEPCFYK--KTCPQVHFIVFKVVEKVSRTDPRM 245
Query: 66 TAKLLRLVYSDCFITGCDASILLDEGPN--TEKKA-PQNRGLGAFVLIDNIKTFVERQCP 122
A L+RL + DCF+ GCDASILL+ +E++A P N + +++ IKT +E+ CP
Sbjct: 246 PASLVRLFFHDCFVQGCDASILLNNTATIVSEQQALPNNNSIRGLDVVNQIKTELEKACP 425
Query: 123 GVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDA--ASVDIPSPSVSWQEALAYF 180
GVVSCADIL LA + LA GP GR+D + ++ A+ ++P+P + + A F
Sbjct: 426 GVVSCADILTLAAEVSSVLAHGPYLKFPLGRRDSLTANRTLANQNLPAPFFNLTQLKAAF 605
Query: 181 KSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRK 240
+GL+ D+ L GAH+ GR C +I DRLYN++GTG DP+++ +L +R++CP +
Sbjct: 606 AVQGLDTTDLVALSGAHSFGRVRCLFILDRLYNFSGTGRPDPTLDTTYLKQLRQICP--Q 779
Query: 241 KGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN--GDDTKEITEEFAAGL 298
G + LV +P + + ++YY + + +L DQ+L + G DT I +F++
Sbjct: 780 GGPPNNLVNFDPTTPDT--LDKNYYSNLQVKKGLLQSDQELFSTPGADTISIVNKFSSDQ 953
Query: 299 QDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFINK 334
F KSF+ SM MGNI VLTG +G+IR+ C F+NK
Sbjct: 954 IAFFKSFSASMIKMGNIGVLTGKKGEIRKQCNFVNK 1061
>TC204493 UP|O22443 (O22443) Seed coat peroxidase precursor , complete
Length = 1294
Score = 204 bits (518), Expect = 6e-53
Identities = 110/279 (39%), Positives = 158/279 (56%), Gaps = 7/279 (2%)
Frame = +1
Query: 62 DKSITAKLLRLVYSDCFITGCDASILLDEGPNTEKKA---PQNRGLGAFVLIDNIKTFVE 118
D I A L+RL + DCF+ GCD S+LL+ E + P + ++++IKT VE
Sbjct: 181 DPRIGASLMRLHFHDCFVQGCDGSVLLNNTDTIESEQDALPNINSIRGLDVVNDIKTAVE 360
Query: 119 RQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDA--ASVDIPSPSVSWQEA 176
CP VSCADIL +A A L GGPG+PV GR+D + ++ A+ ++P+P + +
Sbjct: 361 NSCPDTVSCADILAIAAEIASVLGGGPGWPVPLGRRDSLTANRTLANQNLPAPFFNLTQL 540
Query: 177 LAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLC 236
A F +GLN LD+ TL G HT GR CS +RLYN++ TG+ DP++N +L+ +R C
Sbjct: 541 KASFAVQGLNTLDLVTLSGGHTFGRARCSTFINRLYNFSNTGNPDPTLNTTYLEVLRARC 720
Query: 237 PPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN--GDDTKEITEEF 294
P G + N D + +F YY +L +L DQ+L + G DT I F
Sbjct: 721 PQNATGDN----LTNLDLSTPDQFDNRYYSNLLQLNGLLQSDQELFSTPGADTIPIVNSF 888
Query: 295 AAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
++ F +F VSM MGNI VLTG++G+IR C F+N
Sbjct: 889 SSNQNTFFSNFRVSMIKMGNIGVLTGDEGEIRLQCNFVN 1005
>TC209015 similar to UP|PE72_ARATH (Q9FJZ9) Peroxidase 72 precursor (Atperox
P72) (PRXR8) (ATP6a) , partial (81%)
Length = 1099
Score = 204 bits (518), Expect = 6e-53
Identities = 116/275 (42%), Positives = 164/275 (59%), Gaps = 6/275 (2%)
Frame = +3
Query: 65 ITAKLLRLVYSDCFITGCDASILLD--EGPNTEKKAPQNRGLG-AFVLIDNIKTFVERQC 121
+ A +LRL + DCF+ GCDAS+LLD E N+EK + NR F +ID IK +ERQC
Sbjct: 12 LAASILRLHFHDCFVKGCDASLLLDSSESINSEKGSNPNRNSARGFEVIDAIKAELERQC 191
Query: 122 PGVVSCADILQLATRDAVQLAGGPGYPVFTGRKD--GMRSDAASVDIPSPSVSWQEALAY 179
P VSCADIL LA RD+V L GGP + V GR+D G ++ +IP+P+ ++Q L
Sbjct: 192 PSTVSCADILTLAARDSVVLTGGPNWEVPLGRRDSLGASISGSNNNIPAPNNTFQTILTK 371
Query: 180 FKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPR 239
FK +GL+++D+ L G HTIG C+ RLYN +G G D +++ + T+R CP
Sbjct: 372 FKLQGLDLVDLVALSGGHTIGNARCTTFRQRLYNQSGNGEPDSTLDQYYASTLRTRCP-- 545
Query: 240 KKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLN-GDDTKEITEEFAAGL 298
G L +L D + YKF SY+K +L ++ +L DQ L ++ E+ + +A
Sbjct: 546 SSGGDQNLFFL--DYATPYKFDNSYFKNLLAYKGLLSSDQVLFTMNQESAELVKLYAERN 719
Query: 299 QDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
F + FA SM MGNI LT ++G+IR+NCR IN
Sbjct: 720 DIFFEHFAKSMIKMGNISPLTNSRGEIRENCRRIN 824
>TC204669 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (81%)
Length = 1413
Score = 203 bits (517), Expect = 8e-53
Identities = 118/291 (40%), Positives = 168/291 (57%), Gaps = 1/291 (0%)
Frame = +2
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGPNTEKKAPQNRG 103
C AE VR+ V+ + D ++ +LR+ + DCF+ GCDAS+L+ G TE+ A N
Sbjct: 272 CPRAESIVRSTVESHLRSDPTLAGPILRMHFHDCFVRGCDASVLI-AGAGTERTAGPNLS 448
Query: 104 LGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRS-DAA 162
L F +ID+ K +E CPGVVSCADIL LA RD+V L+GG + V TGRKDG S +
Sbjct: 449 LRGFDVIDDAKAKIEALCPGVVSCADILSLAARDSVVLSGGLSWQVPTGRKDGRVSIGSE 628
Query: 163 SVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDP 222
++ +P P+ + F ++GLN D+ L G HTIG + C DR+YN NGT DP
Sbjct: 629 ALTLPGPNDTVATQKDKFSNKGLNTEDLVILAGGHTIGTSACRSFADRIYNPNGT---DP 799
Query: 223 SMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLL 282
S++ +FL +R++CP Q+ P + D+GS +KF SY+ ++ +L DQ L
Sbjct: 800 SIDPSFLPFLRQICP-----QTQPTKRVALDTGSQFKFDTSYFAHLVRGRGILRSDQVLW 964
Query: 283 NGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
T+ +++ A FK F SM M NI V TG+QG+IR+ C IN
Sbjct: 965 TDASTRGFVQKYLA-TGPFKVQFGKSMIKMSNIGVKTGSQGEIRKICSAIN 1114
>TC215016 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
complete
Length = 1453
Score = 203 bits (516), Expect = 1e-52
Identities = 123/337 (36%), Positives = 184/337 (54%), Gaps = 6/337 (1%)
Frame = +3
Query: 3 SSMCVAFLLVVA-FAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKF 61
S +C +++ + + F LS +A +L+ +Y C AE + V
Sbjct: 126 SRICASYMKMGSNFRFLSLCLLALIASTHAQLQLGFYAKS--CPKAEQIILKFVHEHIHN 299
Query: 62 DKSITAKLLRLVYSDCFITGCDASILLDEGPN-TEKKAPQNRGLGAFVLIDNIKTFVERQ 120
S+ A L+R+ + DCF+ GCDAS+LL+ N EK AP N + F ID IK+ VE +
Sbjct: 300 APSLAAALIRMHFHDCFVRGCDASVLLNSTTNQAEKNAPPNLTVRGFDFIDRIKSLVEAE 479
Query: 121 CPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD--AASVDIPSPSVSWQEALA 178
CPGVVSCADIL L+ RD + GGP + V TGR+DG+ S+ A +IP+PS ++
Sbjct: 480 CPGVVSCADILTLSARDTIVATGGPFWKVPTGRRDGVISNLTEARDNIPAPSSNFTTLQT 659
Query: 179 YFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKL-CP 237
F ++GL++ D+ L GAHTIG HCS +++RL+N+ G G DPS+++ + ++ C
Sbjct: 660 LFANQGLDLKDLVLLSGAHTIGIAHCSSLSNRLFNFTGKGDQDPSLDSEYAANLKAFKCT 839
Query: 238 PRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTK-EITEEFAA 296
K + + D GS F SYY ++ + D LL TK +I E
Sbjct: 840 DLNKLNT---TKIEMDPGSRKTFDLSYYSHVIKRRGLFESDAALLTNSVTKAQIIELLEG 1010
Query: 297 GLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
+++F FA SM MG I V TG +G+IR++C F+N
Sbjct: 1011SVENFFAEFATSMEKMGRINVKTGTEGEIRKHCAFVN 1121
>TC205794 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1205
Score = 203 bits (516), Expect = 1e-52
Identities = 117/309 (37%), Positives = 172/309 (54%), Gaps = 7/309 (2%)
Frame = +2
Query: 32 KLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEG 91
+L+ +Y+ + C VR V+ K D + A L+RL + DCF+ GCDASILL+
Sbjct: 113 QLDPSFYR--DTCPKVHSIVREVVRNVSKSDPRMLASLIRLHFHDCFVQGCDASILLNNT 286
Query: 92 PNTEKKA---PQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYP 148
E + P N + +++ IKT VE CPGVVSCADIL LA + L GP +
Sbjct: 287 ATIESEQQAFPNNNSIRGLDVVNQIKTAVENACPGVVSCADILALAAEISSVLGHGPDWK 466
Query: 149 VFTGRKDGMRSDA--ASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSY 206
V GR+D + ++ A+ ++P+P ++ + F +GLN D+ L GAHTIGR C +
Sbjct: 467 VPLGRRDSLTANRTLANQNLPAPFLNLTQLKDAFAVQGLNTTDLVALSGAHTIGRAQCRF 646
Query: 207 ITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYK 266
DRLYN++ TG+ DP++N +L T+ +CP G + N D + +YY
Sbjct: 647 FVDRLYNFSSTGNPDPTLNTTYLQTLSAICPNGGPGTN----LTNFDPTTPDTVDSNYYS 814
Query: 267 RILNHEAVLGIDQQLLN--GDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQ 324
+ ++ +L DQ+L + G DT I F++ F ++F SM MGNI VLTG+QG+
Sbjct: 815 NLQVNKGLLQSDQELFSTTGTDTIAIVNSFSSNQTLFFENFKASMIKMGNIGVLTGSQGE 994
Query: 325 IRQNCRFIN 333
IRQ C FIN
Sbjct: 995 IRQQCNFIN 1021
>TC215014 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
complete
Length = 1341
Score = 198 bits (504), Expect = 2e-51
Identities = 110/275 (40%), Positives = 160/275 (58%), Gaps = 5/275 (1%)
Frame = +1
Query: 64 SITAKLLRLVYSDCFITGCDASILLDEGPN-TEKKAPQNRGLGAFVLIDNIKTFVERQCP 122
S+ A L+R+ + DCF+ GCD S+LL+ N EK AP N + F ID IK+ VE +CP
Sbjct: 250 SLAAALIRMHFHDCFVRGCDGSVLLNSTTNQAEKNAPPNLTVRGFDFIDRIKSLVEAECP 429
Query: 123 GVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD--AASVDIPSPSVSWQEALAYF 180
GVVSCADIL LA RD + GGP + V TGR+DG+ S+ A +IP+PS ++ F
Sbjct: 430 GVVSCADILTLAARDTIVATGGPFWKVPTGRRDGVVSNLTEARNNIPAPSSNFTTLQTLF 609
Query: 181 KSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKL-CPPR 239
++GL++ D+ L GAHTIG HCS +++RL+N+ G G DPS+++ + ++ C
Sbjct: 610 ANQGLDLKDLVLLSGAHTIGIAHCSSLSNRLFNFTGKGDQDPSLDSEYAANLKAFKCTDL 789
Query: 240 KKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTK-EITEEFAAGL 298
K + + D GS F SYY ++ + D LL TK +I + +
Sbjct: 790 NKLNT---TKIEMDPGSRKTFDLSYYSHVIKRRGLFESDAALLTNSVTKAQIIQLLEGSV 960
Query: 299 QDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
++F FA S+ MG I V TG +G+IR++C FIN
Sbjct: 961 ENFFAEFATSIEKMGRINVKTGTEGEIRKHCAFIN 1065
>TC207747 weakly similar to UP|Q9M4Z5 (Q9M4Z5) Peroxidase prx12 precursor ,
partial (88%)
Length = 1204
Score = 197 bits (502), Expect = 4e-51
Identities = 115/295 (38%), Positives = 167/295 (55%), Gaps = 5/295 (1%)
Frame = +3
Query: 44 CRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEGP--NTEKKAPQN 101
C AE V+++V+ + I A L+R+ + DCFI GCDAS+LLD P EK +P N
Sbjct: 150 CSMAEFIVKDEVRKGVTNNPGIAAGLVRMHFHDCFIRGCDASVLLDSTPLNTAEKDSPAN 329
Query: 102 R-GLGAFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSD 160
+ L + +IDN K +E CPG+VSCADI+ A RD+V+ A G GY V GR+DG S
Sbjct: 330 KPSLRGYEVIDNAKAKLEAVCPGIVSCADIVAFAARDSVEFARGLGYDVPAGRRDGRISL 509
Query: 161 AASV--DIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTG 218
A+ ++P P+ + + F +GL +M TL GAHTIGR+HCS + RLYN++ T
Sbjct: 510 ASDTRTELPPPTFNVNQLTQLFARKGLTQDEMVTLSGAHTIGRSHCSAFSSRLYNFSTTS 689
Query: 219 SSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGID 278
S DPS++ ++ +++ CP +G ++ + + D S YY IL + + D
Sbjct: 690 SQDPSLDPSYAALLKRQCP---QGSTNQNLVVPMDPSSPGIADVGYYVDILANRGLFTSD 860
Query: 279 QQLLNGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
Q LL +T ++ A + FA +M MG I VL GN G+IR NCR +N
Sbjct: 861 QTLLTNAETASQVKQNARDPYLWASQFADAMVKMGQIIVLKGNAGEIRTNCRVVN 1025
>TC230907 similar to UP|O24081 (O24081) Peroxidase1A precursor , partial
(88%)
Length = 944
Score = 196 bits (497), Expect = 2e-50
Identities = 106/279 (37%), Positives = 159/279 (55%), Gaps = 7/279 (2%)
Frame = +2
Query: 62 DKSITAKLLRLVYSDCFITGCDASILLDEGP---NTEKKAPQNRGLGAFVLIDNIKTFVE 118
D I A L+RL + DCF+ GCDASILL++ + + P N + +++ IKT VE
Sbjct: 41 DPRILASLIRLHFHDCFVQGCDASILLNDTDTIVSEQSAVPNNNSIRGLDVVNQIKTAVE 220
Query: 119 RQCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDA--ASVDIPSPSVSWQEA 176
CPG+VSCADIL LA + + LA GP + V GR+D + ++ A+ ++P+P+ + +
Sbjct: 221 NACPGIVSCADILALAAQISSDLANGPVWQVPLGRRDSLTANQTLANQNLPAPTFTIDQL 400
Query: 177 LAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLC 236
+ F ++ LN+ D+ L GAHTIGR C + DRLYN++ TG+ DP++N L +++ +C
Sbjct: 401 IESFGNQSLNITDLVALSGAHTIGRAQCRFFVDRLYNFSNTGNPDPTLNTTLLQSLQGIC 580
Query: 237 PPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLL--NGDDTKEITEEF 294
P G + N D + F +YY + +L DQ+LL N D I F
Sbjct: 581 PNGGPGTN----LTNLDLTTPDTFDSNYYSNLQLQNGLLQSDQELLSANNTDIVAIVNNF 748
Query: 295 AAGLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
+ F ++F SM MGNI VLTG+QG+IR C +N
Sbjct: 749 ISNQTLFFENFKASMIKMGNIGVLTGSQGEIRSQCNSVN 865
>TC205795 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(96%)
Length = 1321
Score = 194 bits (492), Expect = 6e-50
Identities = 119/324 (36%), Positives = 174/324 (52%), Gaps = 7/324 (2%)
Frame = +2
Query: 17 ALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSD 76
AL V T +L+ +YK + C VR V+ K D I A L+RL + D
Sbjct: 68 ALCCVVVMLLTLSDAQLDNSFYK--DTCPRVHSIVREVVRNVSKSDPRILASLIRLHFHD 241
Query: 77 CFITGCDASILLDEGPN--TEKKAP-QNRGLGAFVLIDNIKTFVERQCPGVVSCADILQL 133
CF+ GCDASILL++ +E+ AP N + +++ IKT VE CPG+VSCADIL L
Sbjct: 242 CFVQGCDASILLNDTATIVSEQSAPPNNNSIRGLDVVNQIKTAVENACPGIVSCADILAL 421
Query: 134 ATRDAVQLAGGPGYPVFTGRKDGMRSD--AASVDIPSPSVSWQEALAYFKSRGLNVLDMG 191
A + LA GP + V GR+D + S A ++P + + + + F +GLN D+
Sbjct: 422 AAEISSVLAHGPDWKVPLGRRDSLNSSFSLALQNLPGFNFTLDQLKSTFDRQGLNTTDLV 601
Query: 192 TLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYLN 251
L GAHTIGR+ C + R+YN++G G+SDP++N +R +CP G + N
Sbjct: 602 ALSGAHTIGRSQCRFFAHRIYNFSGNGNSDPTLNTTLSQALRAICPNGGPGTN----LTN 769
Query: 252 PDSGSSYKFTESYYKRILNHEAVLGIDQQLL--NGDDTKEITEEFAAGLQDFKKSFAVSM 309
D + +F +YY + +L DQ L +G +T I F + F + F VSM
Sbjct: 770 LDLTTPDRFDSNYYSNLQLQNGLLRSDQVLFSTSGAETIAIVNSFGSNQTLFYEHFKVSM 949
Query: 310 YNMGNIKVLTGNQGQIRQNCRFIN 333
M I+VLTG+QG+IR++C F+N
Sbjct: 950 IKMSIIEVLTGSQGEIRKHCNFVN 1021
>TC206181 similar to UP|Q7XYR7 (Q7XYR7) Class III peroxidase, partial (94%)
Length = 1219
Score = 192 bits (487), Expect = 2e-49
Identities = 118/311 (37%), Positives = 172/311 (54%), Gaps = 9/311 (2%)
Frame = +2
Query: 32 KLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVYSDCFITGCDASILLDEG 91
+L ++Y ++ C + V++ V+ + + A LLRL + DCF+ GCD SILLD+
Sbjct: 131 QLSTNFY--YHSCPNLFSSVKSTVQSAISKETRMGASLLRLFFHDCFVNGCDGSILLDDT 304
Query: 92 PNT--EKKAPQNRGLG-AFVLIDNIKTFVERQCPGVVSCADILQLATRDAVQLAGGPGYP 148
+ EK A NR F +IDNIK+ VE+ CPGVVSCADIL +A RD+VQ+ GGP +
Sbjct: 305 SSFTGEKNANPNRNSARGFEVIDNIKSAVEKVCPGVVSCADILAIAARDSVQILGGPSWN 484
Query: 149 VFTGRKDGMRS--DAASVDIPSPSVSWQEALAYFKSRGLNVLDMGTLLGAHTIGRTHCSY 206
V GR+D + AA+ IP P+ + + ++ F + GL+ D+ L G HTIG+ C+
Sbjct: 485 VKLGRRDARTASQSAANNGIPPPTSNLNQLISRFSALGLSTKDLVALSGGHTIGQARCTN 664
Query: 207 ITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSD----PLVYLNPDSGSSYKFTE 262
R+YN + ++ AF T ++ C PR G D PL P S +Y
Sbjct: 665 FRARIYN-------ETNIETAFARTRQQSC-PRTSGSGDNNLAPLDLQTPTSFDNY---- 808
Query: 263 SYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKKSFAVSMYNMGNIKVLTGNQ 322
Y+K ++ + +L DQQL NG T I ++ F FA +M MG+I LTG+
Sbjct: 809 -YFKNLVQKKGLLHSDQQLFNGGSTDSIVRGYSTNPGTFSSDFAAAMIKMGDISPLTGSN 985
Query: 323 GQIRQNCRFIN 333
G+IR+NCR IN
Sbjct: 986 GEIRKNCRRIN 1018
>TC216324 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, complete
Length = 1246
Score = 190 bits (483), Expect = 7e-49
Identities = 106/268 (39%), Positives = 150/268 (55%), Gaps = 4/268 (1%)
Frame = +2
Query: 67 AKLLRLVYSDCFITGCDASILLDEGPNTEKKAP-QNRGLGAFVLIDNIKTFVERQCPGVV 125
A ++RL + DCF+ GCD S+LLD GP++EK AP N L + +ID IK+ VE CPGVV
Sbjct: 224 ASIVRLFFHDCFVNGCDGSVLLD-GPSSEKTAPPNNNSLRGYEVIDAIKSKVETVCPGVV 400
Query: 126 SCADILQLATRDAVQLAGGPGYPVFTGRKD---GMRSDAASVDIPSPSVSWQEALAYFKS 182
SCADI+ +A RD+V + GGP + V GR+D G + A S +P P+ S + F
Sbjct: 401 SCADIVTIAARDSVAILGGPNWKVKLGRRDSTTGFFNLANSGVLPGPNSSLSSLIQRFDD 580
Query: 183 RGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKG 242
+GL+ DM L GAHTIG+ C DR+YN N ++++ F +K CP G
Sbjct: 581 QGLSTKDMVALSGAHTIGKARCVSYRDRIYNEN-------NIDSLFAKARQKNCPKGSSG 739
Query: 243 QSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFK 302
D + F Y+K ++N + +L DQ+L NG T + ++ + F+
Sbjct: 740 TPKDNNVAPLDFKTPNHFDNEYFKNLINKKGLLRSDQELFNGGSTDSLVRTYSNNQRVFE 919
Query: 303 KSFAVSMYNMGNIKVLTGNQGQIRQNCR 330
F +M MGNIK LTG+ GQIR+ CR
Sbjct: 920 ADFVTAMIKMGNIKPLTGSNGQIRKQCR 1003
>TC216945 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (96%)
Length = 1274
Score = 189 bits (481), Expect = 1e-48
Identities = 123/337 (36%), Positives = 177/337 (52%), Gaps = 11/337 (3%)
Frame = +2
Query: 8 AFLLVVAFAALSSSPV--AAATPPRPK-LEWHYYKTHNICRDAELYVRNQVKLFWKFDKS 64
A LL+ F ++ + V A A PP K L + +Y C + VR+++K + D +
Sbjct: 26 ALLLISIFLSVYNIEVCEAQARPPTAKGLSYTFYDKS--CPKLKSIVRSELKKVFNKDIA 199
Query: 65 ITAKLLRLVYSDCFITGCDASILLD---EGPNTEKKAPQNRGLG--AFVLIDNIKTFVER 119
A LLRL + DCF+ GCD S+LLD GP EK+AP N L AF +I+N++ +E+
Sbjct: 200 QAAGLLRLHFHDCFVQGCDGSVLLDGSASGPG-EKEAPPNLTLRPEAFKIIENLRGLLEK 376
Query: 120 QCPGVVSCADILQLATRDAVQLAGGPGYPVFTGRKDGMRSDAASV---DIPSPSVSWQEA 176
C VVSC+DI L RDAV L+GGP Y + GR+DG+ V ++P PS +
Sbjct: 377 SCGRVVSCSDITALTARDAVFLSGGPDYEIPLGRRDGLTFATRQVTLDNLPPPSSNASTI 556
Query: 177 LAYFKSRGLNVLDMGTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLC 236
L+ ++ L+ D+ L G HTIG +HC T+RLY + DP M+ F + +R+ C
Sbjct: 557 LSSLATKNLDPTDVVALSGGHTIGISHCGSFTNRLYP-----TQDPVMDKTFGNNLRRTC 721
Query: 237 PPRKKGQSDPLVYLNPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAA 296
P + L +P++ F YY ++N + + DQ L TK I +FA
Sbjct: 722 PAANTDNTTVLDIRSPNT-----FDNKYYVDLMNRQGLFTSDQDLYTNTRTKGIVTDFAV 886
Query: 297 GLQDFKKSFAVSMYNMGNIKVLTGNQGQIRQNCRFIN 333
F F +M MG + VLTGNQG+IR NC N
Sbjct: 887 NQSLFFDKFVFAMLKMGQLNVLTGNQGEIRANCSVRN 997
>TC226717 homologue to UP|Q9ZTW8 (Q9ZTW8) Peroxidase, partial (85%)
Length = 1343
Score = 189 bits (480), Expect = 2e-48
Identities = 118/323 (36%), Positives = 168/323 (51%), Gaps = 4/323 (1%)
Frame = +3
Query: 15 FAALSSSPVAAATPPRPKLEWHYYKTHNICRDAELYVRNQVKLFWKFDKSITAKLLRLVY 74
F AL +A + +L+ +Y C AE + V S+ A L+RL +
Sbjct: 108 FKALIICLIALIGSTQAQLQLGFYAKS--CPKAEQIILKYVVEHIHNAPSLAAALIRLHF 281
Query: 75 SDCFITGCDASILLDEGPNT--EKKAPQNRGLGAFVLIDNIKTFVERQCPGVVSCADILQ 132
DCF+ GCD S+L+D P EK A N L F I+ IK VE +CPGVVSCADIL
Sbjct: 282 HDCFVNGCDGSVLVDSTPGNQAEKDAIPNLTLRGFGFIEAIKRLVEAECPGVVSCADILA 461
Query: 133 LATRDAVQLAGGPGYPVFTGRKDGMRSDAASV--DIPSPSVSWQEALAYFKSRGLNVLDM 190
L RD++ GGP + V TGR+DG S AA +P+P + L F + GL+ D+
Sbjct: 462 LTARDSIHATGGPYWNVPTGRRDGFISRAADPLRSLPAPFHNLTTQLTLFGNVGLDANDL 641
Query: 191 GTLLGAHTIGRTHCSYITDRLYNYNGTGSSDPSMNAAFLDTMRKLCPPRKKGQSDPLVYL 250
L+GAHTIG HCS I+ RLYN+ G G +DP+++ + ++ K + L+ +
Sbjct: 642 VLLVGAHTIGIAHCSSISTRLYNFTGKGDTDPTIDNGYAKNLKTF--KCKNINDNSLIEM 815
Query: 251 NPDSGSSYKFTESYYKRILNHEAVLGIDQQLLNGDDTKEITEEFAAGLQDFKKSFAVSMY 310
+P GS F YYK+++ + D +LL T+ I Q F FA SM
Sbjct: 816 DP--GSRDTFDLGYYKQVVKRRGLFQSDAELLTSPITRSIIASQLQSTQGFFAEFAKSME 989
Query: 311 NMGNIKVLTGNQGQIRQNCRFIN 333
MG I V G++G+IR++C +N
Sbjct: 990 KMGRINVKLGSEGEIRKHCARVN 1058
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,749,361
Number of Sequences: 63676
Number of extensions: 198297
Number of successful extensions: 1434
Number of sequences better than 10.0: 191
Number of HSP's better than 10.0 without gapping: 1225
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1232
length of query: 334
length of database: 12,639,632
effective HSP length: 98
effective length of query: 236
effective length of database: 6,399,384
effective search space: 1510254624
effective search space used: 1510254624
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0106.6


