

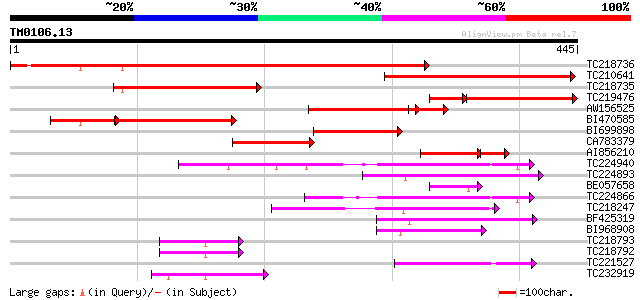
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.13
(445 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218736 similar to UP|CV4L_HUMAN (Q9NU19) TBC1 domain family pr... 523 e-149
TC210641 weakly similar to UP|Q6PUR3 (Q6PUR3) C22orf4-like prote... 291 5e-79
TC218735 177 8e-45
TC219476 174 9e-44
AW156525 127 3e-38
BI470585 149 2e-36
BI699898 132 4e-31
CA783379 125 3e-29
AI856210 80 1e-22
TC224940 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein... 79 4e-15
TC224893 67 2e-11
BE057658 62 7e-10
TC224866 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein... 61 1e-09
TC218247 similar to UP|Q9FFV0 (Q9FFV0) Similarity to GTPase acti... 57 2e-08
BF425319 55 5e-08
BI968908 52 4e-07
TC218793 weakly similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial... 52 7e-07
TC218792 homologue to UP|O44991 (O44991) Lipid depleted protein ... 49 3e-06
TC221527 similar to UP|O81347 (O81347) Plant adhesion molecule 1... 46 4e-05
TC232919 44 1e-04
>TC218736 similar to UP|CV4L_HUMAN (Q9NU19) TBC1 domain family protein
C6orf197, partial (23%)
Length = 1212
Score = 523 bits (1346), Expect = e-149
Identities = 267/334 (79%), Positives = 286/334 (84%), Gaps = 5/334 (1%)
Frame = +2
Query: 1 MKNNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSN--LSSP 58
M ED N DSRF+QTLR+VQGLLKGRS+PGKILLS+RVDP DNSN +SSP
Sbjct: 65 MNKRQEDLRNIATT--LDSRFNQTLRNVQGLLKGRSIPGKILLSRRVDPPDNSNSKISSP 238
Query: 59 TYTRSNSYNDAGTSDHASETVEVEVHSSS---GIPGENKLKISTSHVENPSEDVRKSSMG 115
Y RS S+NDAGTSD+ S VE E S S I NKLK+STS +P E+ KS+MG
Sbjct: 239 NYKRSFSHNDAGTSDNTSGAVEEEFQSKSKPISIANANKLKVSTSLGGSPPEEFHKSTMG 418
Query: 116 ARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGV 175
ARATDSAR+MKFTKVLSGTVVILDKLRELAWSGVPD MRP VWRLLLGYAPPNSDRREGV
Sbjct: 419 ARATDSARIMKFTKVLSGTVVILDKLRELAWSGVPDNMRPKVWRLLLGYAPPNSDRREGV 598
Query: 176 LRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERI 235
LRRKRLEYLDC+SQYYDIPDTERS+DE+NML QI +DCPRTVPDV FFQQQQVQKSLERI
Sbjct: 599 LRRKRLEYLDCISQYYDIPDTERSDDEVNMLHQIGIDCPRTVPDVPFFQQQQVQKSLERI 778
Query: 236 LYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWC 295
LYAWAIRHPASGYVQGINDLVTPF VVFLSE+ EG I+NWSMSDLSSD ISN+EADCYWC
Sbjct: 779 LYAWAIRHPASGYVQGINDLVTPFLVVFLSEHFEGDINNWSMSDLSSDIISNIEADCYWC 958
Query: 296 LSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRID 329
LSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRID
Sbjct: 959 LSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRID 1060
>TC210641 weakly similar to UP|Q6PUR3 (Q6PUR3) C22orf4-like protein, partial
(14%)
Length = 707
Score = 291 bits (744), Expect = 5e-79
Identities = 138/150 (92%), Positives = 144/150 (96%)
Frame = +1
Query: 295 CLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCL 354
CLSKLLD MQDHYTFAQPGIQRLVFKLKELVRRIDDPVS HME QGLEFLQFAFRWFNCL
Sbjct: 1 CLSKLLDSMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSNHMEEQGLEFLQFAFRWFNCL 180
Query: 355 LIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPT 414
LIREIPF++VTRLWDTYLAEGDALPDFLVYI ASFLLTWSD LQKLDFQ+LVMFLQHLPT
Sbjct: 181 LIREIPFHLVTRLWDTYLAEGDALPDFLVYISASFLLTWSDNLQKLDFQELVMFLQHLPT 360
Query: 415 QDWTHQELEMVLSRAFMWHSMFNNSPNHLA 444
++WT QELEMVLSRAFMWHSMFNNSP+HLA
Sbjct: 361 KNWTDQELEMVLSRAFMWHSMFNNSPSHLA 450
>TC218735
Length = 366
Score = 177 bits (449), Expect = 8e-45
Identities = 94/121 (77%), Positives = 103/121 (84%), Gaps = 5/121 (4%)
Frame = +3
Query: 82 EVHSSS---GIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVIL 138
EVHS+S GIP ENKLKIS S+VE+ SE++RKSSMGARATDSARVMKFTKVLS T+V L
Sbjct: 3 EVHSTSKPFGIPNENKLKISASNVESSSEELRKSSMGARATDSARVMKFTKVLSETMVKL 182
Query: 139 DKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLD-CVSQYYD-IPDT 196
+KLRE +W GVPDYMRPTVWRLLLGYAPPNSDRREGVL+RKRLEYLD C D PDT
Sbjct: 183 EKLREFSWRGVPDYMRPTVWRLLLGYAPPNSDRREGVLKRKRLEYLDWCFFSIMDNFPDT 362
Query: 197 E 197
E
Sbjct: 363 E 365
>TC219476
Length = 899
Score = 174 bits (440), Expect = 9e-44
Identities = 78/87 (89%), Positives = 87/87 (99%)
Frame = +2
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWT 418
IPF+++TRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQ+LVMFLQHLPT++WT
Sbjct: 374 IPFHLITRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQELVMFLQHLPTENWT 553
Query: 419 HQELEMVLSRAFMWHSMFNNSPNHLAT 445
HQELEMVLSRAFMWH+MFNNSP+HLA+
Sbjct: 554 HQELEMVLSRAFMWHTMFNNSPSHLAS 634
Score = 60.1 bits (144), Expect = 2e-09
Identities = 25/30 (83%), Positives = 29/30 (96%)
Frame = +1
Query: 330 DPVSTHMENQGLEFLQFAFRWFNCLLIREI 359
+PVS H+E+QGLEFLQFAFRWFNCLLIRE+
Sbjct: 136 EPVSRHIEDQGLEFLQFAFRWFNCLLIREV 225
>AW156525
Length = 338
Score = 127 bits (319), Expect(2) = 3e-38
Identities = 64/88 (72%), Positives = 68/88 (76%)
Frame = +3
Query: 235 ILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYW 294
IL WAIRH ASGYV GINDLVTPF VVFLS+Y G IDNWSMSDLSS EISNV ADCYW
Sbjct: 6 ILNTWAIRHAASGYVHGINDLVTPFLVVFLSQY**GGIDNWSMSDLSSHEISNV*ADCYW 185
Query: 295 CLSKLLDGMQDHYTFAQPGIQRLVFKLK 322
LSKLLD +QDHYT AQ + F L+
Sbjct: 186 GLSKLLDSLQDHYTCAQHSTPKACF*LE 269
Score = 49.7 bits (117), Expect(2) = 3e-38
Identities = 23/31 (74%), Positives = 25/31 (80%)
Frame = +1
Query: 314 IQRLVFKLKELVRRIDDPVSTHMENQGLEFL 344
++RL F KELV RIDDPVS HME QGLEFL
Sbjct: 244 LRRLAFNSKELVMRIDDPVSNHMEEQGLEFL 336
>BI470585
Length = 422
Score = 149 bits (376), Expect = 2e-36
Identities = 77/96 (80%), Positives = 82/96 (85%)
Frame = +3
Query: 83 VHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLR 142
+ S G E KLK+STS +P E+ RKS+MGARATDSARVMKFTKVLSGTVVILDKLR
Sbjct: 135 IWGSRGGISEYKLKVSTSLGGSPPEEFRKSTMGARATDSARVMKFTKVLSGTVVILDKLR 314
Query: 143 ELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
ELAWSGVPD MRP VWRLLLGYAPPNSDRREGVLRR
Sbjct: 315 ELAWSGVPDNMRPKVWRLLLGYAPPNSDRREGVLRR 422
Score = 75.1 bits (183), Expect = 6e-14
Identities = 40/57 (70%), Positives = 44/57 (77%), Gaps = 2/57 (3%)
Frame = +1
Query: 33 KGRSMPGKILLSQRVDPIDNSN--LSSPTYTRSNSYNDAGTSDHASETVEVEVHSSS 87
KGRS+PGKILLS+RVDP DNSN SSP Y RS S+NDAGTSDH S VE E S+S
Sbjct: 1 KGRSIPGKILLSRRVDPPDNSNSKASSPNYKRSFSHNDAGTSDHTSGAVEEEFQSTS 171
>BI699898
Length = 421
Score = 132 bits (331), Expect = 4e-31
Identities = 60/70 (85%), Positives = 62/70 (87%)
Frame = +3
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVT F VVF EY EG IDNWSMSDLSSD ISN+E DCYWCLSK
Sbjct: 210 WAIRHPASGYVQGINDLVTLFLVVF*LEYFEGDIDNWSMSDLSSDVISNIEVDCYWCLSK 389
Query: 299 LLDGMQDHYT 308
L+DGMQDHYT
Sbjct: 390 LIDGMQDHYT 419
>CA783379
Length = 455
Score = 125 bits (315), Expect = 3e-29
Identities = 59/64 (92%), Positives = 62/64 (96%)
Frame = +3
Query: 176 LRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERI 235
L+RK LEYLDCVSQYYDIPDTERS+DEINMLRQIAVDCPRTVP+VSFFQQQQVQKSLERI
Sbjct: 3 LKRKPLEYLDCVSQYYDIPDTERSDDEINMLRQIAVDCPRTVPEVSFFQQQQVQKSLERI 182
Query: 236 LYAW 239
LY W
Sbjct: 183 LYTW 194
>AI856210
Length = 464
Score = 80.1 bits (196), Expect(2) = 1e-22
Identities = 36/49 (73%), Positives = 43/49 (87%)
Frame = +1
Query: 323 ELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTY 371
EL+RRID+PVS H+E+Q LEFLQFAF WFN L+I EIPF +VTRLWDT+
Sbjct: 1 ELIRRIDEPVSWHIEDQRLEFLQFAFCWFNYLVIHEIPFLLVTRLWDTF 147
Score = 44.3 bits (103), Expect(2) = 1e-22
Identities = 21/23 (91%), Positives = 22/23 (95%)
Frame = +2
Query: 370 TYLAEGDALPDFLVYIFASFLLT 392
T+LAEGDAL DFLVYIFASFLLT
Sbjct: 140 THLAEGDALADFLVYIFASFLLT 208
>TC224940 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein-like,
partial (66%)
Length = 1783
Score = 79.0 bits (193), Expect = 4e-15
Identities = 71/289 (24%), Positives = 124/289 (42%), Gaps = 9/289 (3%)
Frame = +2
Query: 133 GTVVILDKLRELAWSGVPDY-MRPTVWRLLLGYAPPNSD--RREGVLRRKRLEYLDCVSQ 189
G V + LR+ + G D+ ++ VW LLLGY P S RE + K+LEY + +Q
Sbjct: 734 GRVTDSEALRKRVFYGGLDHKLQNEVWGLLLGYYPYESTYAEREFLKSVKKLEYENIKNQ 913
Query: 190 YYDIPDTERSEDEINMLRQ--IAVDCPRTVPDVSFFQQQQVQKS--LERILYAWAIRHPA 245
+ I + R+ I D RT ++F++ L IL ++ +
Sbjct: 914 WQSISSAQAKRFTKFRERKGLIEKDVVRTDRSLAFYEGDDNPNVNVLRDILLTYSFYNFD 1093
Query: 246 SGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQD 305
GY QG++DL++P V M D E++ +WC L++ +
Sbjct: 1094LGYCQGMSDLLSPILFV--------------MDD---------ESEAFWCFVALMERLGP 1204
Query: 306 HYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVT 365
++ Q G+ +F L +LV +D P+ + + + F FRW RE +
Sbjct: 1205NFNRDQNGMHSQLFALSKLVELLDSPLHNYFKQRDCLNYFFCFRWILIQFKREFEYEKTM 1384
Query: 366 RLWDTYLAEGDALPDFLVYIFASFLLTWSDKL--QKLDFQDLVMFLQHL 412
RLW+ + +Y+ + L + K+ +++DF L+ F+ L
Sbjct: 1385RLWEVLWTHYPS-EHLHLYVCVAILKRYRGKIIGEEMDFDTLLKFINEL 1528
>TC224893
Length = 705
Score = 66.6 bits (161), Expect = 2e-11
Identities = 42/147 (28%), Positives = 73/147 (49%), Gaps = 5/147 (3%)
Frame = +1
Query: 278 SDLSSDEISNVEADCYWCLSKLLDGMQDHYTF----AQPGIQRLVFKLKELVRRIDDPVS 333
+D + + EAD +C +LL G +D++ + GI+ + +L +L+R D+ +
Sbjct: 10 NDPDEENAAFAEADAVFCFVELLSGFRDNFVQQLDNSVVGIRATITRLSQLLREHDEELW 189
Query: 334 THMENQGLEFLQF-AFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLT 392
H+E QF AFRW LL +E F +WDT L++ D + L+ + + L+
Sbjct: 190 RHLEVTSKVNPQFYAFRWITLLLTQEFNFADSLHIWDTLLSDPDGPQETLLRVCCAMLVL 369
Query: 393 WSDKLQKLDFQDLVMFLQHLPTQDWTH 419
+L DF + LQ+ PT + +H
Sbjct: 370 VRKRLLAGDFTSNLKLLQNYPTTNISH 450
>BE057658
Length = 375
Score = 61.6 bits (148), Expect = 7e-10
Identities = 35/90 (38%), Positives = 41/90 (44%), Gaps = 48/90 (53%)
Frame = +3
Query: 330 DPVSTHMENQGLEFLQFAFRWFNCLLIRE------------------------------- 358
+PVS H+E+QGLEFLQFAFRWFNCLLIRE
Sbjct: 105 EPVSRHIEDQGLEFLQFAFRWFNCLLIREVCNDILFSYKGGSFLNI*LIT*QNGNIGLR* 284
Query: 359 -----------------IPFNMVTRLWDTY 371
IPF+++TRLWDTY
Sbjct: 285 IGMIXCFFTNDV*LYN*IPFHLITRLWDTY 374
>TC224866 similar to UP|Q6ZFZ1 (Q6ZFZ1) GTPase activating protein-like,
partial (31%)
Length = 838
Score = 60.8 bits (146), Expect = 1e-09
Identities = 42/183 (22%), Positives = 79/183 (42%), Gaps = 2/183 (1%)
Frame = +3
Query: 232 LERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEAD 291
L IL ++ + GY QG++DL++P V +DN E++
Sbjct: 57 LRDILLTYSFYNFDLGYCQGMSDLLSPILFV---------MDN--------------ESE 167
Query: 292 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWF 351
+WC L++ + ++ Q G+ +F L +LV +D P+ + + + F FRW
Sbjct: 168 AFWCFVALMERLGPNFNRDQNGMHSQLFALSKLVELLDSPLHNYFKQRDCLNYFFCFRWI 347
Query: 352 NCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKL--QKLDFQDLVMFL 409
RE + RLW+ + +Y+ + L + K+ +++DF L+ F+
Sbjct: 348 LIQFKREFEYEKTMRLWEVLWTHYPS-EHLHLYVCVAILKRYRGKIIGEQMDFDTLLKFI 524
Query: 410 QHL 412
L
Sbjct: 525 NEL 533
>TC218247 similar to UP|Q9FFV0 (Q9FFV0) Similarity to GTPase activating
protein, partial (77%)
Length = 1418
Score = 56.6 bits (135), Expect = 2e-08
Identities = 51/183 (27%), Positives = 80/183 (42%), Gaps = 4/183 (2%)
Frame = +2
Query: 206 LRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLS 265
L QI +D RT + F+++Q+ L IL +A GY QG+ DL +P ++
Sbjct: 332 LHQIGLDVVRTDRTLVFYEKQENLSKLWDILAVYAWIDKDVGYGQGMCDLCSPMIILL-- 505
Query: 266 EYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLKE 323
+ EAD +WC +L+ ++ ++ T + G+ + L
Sbjct: 506 ---------------------DDEADAFWCFERLMRRLRGNFRCTESSVGVAAQLSNLAS 622
Query: 324 LVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLA-EGDALPD-F 381
+ + ID + H+E+ G FAFR L RE F LW+ A E D PD F
Sbjct: 623 VTQVIDPKLHKHLEHLGGGDYLFAFRMLMVLFRREFSFCDSLYLWEMMWALEYD--PDLF 796
Query: 382 LVY 384
L+Y
Sbjct: 797 LMY 805
>BF425319
Length = 412
Score = 55.5 bits (132), Expect = 5e-08
Identities = 37/131 (28%), Positives = 64/131 (48%), Gaps = 5/131 (3%)
Frame = +1
Query: 289 EADCYWCLSKLLDGMQDHYTFAQP----GIQRLVFKLKELVRRIDDPVSTHMENQGLEFL 344
EAD ++C +LL QD++ GI+ + +L +L++ D+ + H+E
Sbjct: 7 EADTFFCFVELLSRFQDNFCQQLDNSIVGIRSTITRLSQLLKEHDEELWRHLEVTTKVNP 186
Query: 345 QF-AFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQ 403
QF AFRW LL +E F + +WD L++ + + L+ I + L+ +L DF
Sbjct: 187 QFYAFRWIILLLTQEFNFADILHIWDVILSDPEGPQETLLRICCAMLILVRRRLLAGDFT 366
Query: 404 DLVMFLQHLPT 414
+ LQ P+
Sbjct: 367 SNLKMLQSYPS 399
>BI968908
Length = 394
Score = 52.4 bits (124), Expect = 4e-07
Identities = 31/91 (34%), Positives = 46/91 (50%), Gaps = 5/91 (5%)
Frame = -2
Query: 289 EADCYWCLSKLLDGMQD----HYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFL 344
EAD ++C +LL G +D H + GI+ + +L +L+R D+ + H+E
Sbjct: 390 EADAFFCFVELLSGFRDNFVHHLANSVVGIRXTITRLSQLLRDHDEELWRHLEVTSKVNP 211
Query: 345 QF-AFRWFNCLLIREIPFNMVTRLWDTYLAE 374
QF AFRW LL +E F WDT L +
Sbjct: 210 QFYAFRWITLLLTQEFNFAATLXXWDTLLTD 118
>TC218793 weakly similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial (12%)
Length = 632
Score = 51.6 bits (122), Expect = 7e-07
Identities = 29/68 (42%), Positives = 39/68 (56%), Gaps = 2/68 (2%)
Frame = +3
Query: 118 ATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDY--MRPTVWRLLLGYAPPNSDRREGV 175
A D +R + LS VV + +LR LA G+PD +R TVW+LLLGY PP+
Sbjct: 393 AHDVSRQAQLLAELSKKVVDMSELRSLACQGIPDAAGIRSTVWKLLLGYLPPDRGLWSAE 572
Query: 176 LRRKRLEY 183
L +KR +Y
Sbjct: 573 LAKKRFQY 596
>TC218792 homologue to UP|O44991 (O44991) Lipid depleted protein 6, partial
(4%)
Length = 756
Score = 49.3 bits (116), Expect = 3e-06
Identities = 28/68 (41%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Frame = +3
Query: 118 ATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDY--MRPTVWRLLLGYAPPNSDRREGV 175
A D +R + LS VV + +LR LA G+PD +R T W+LLLGY PP+
Sbjct: 318 AHDVSRQAQLLAELSKKVVDMSELRSLACQGIPDAAGIRSTAWKLLLGYLPPDRGLWSAE 497
Query: 176 LRRKRLEY 183
L +KR +Y
Sbjct: 498 LAKKRSQY 521
>TC221527 similar to UP|O81347 (O81347) Plant adhesion molecule 1 (PAM1),
partial (41%)
Length = 638
Score = 45.8 bits (107), Expect = 4e-05
Identities = 28/111 (25%), Positives = 52/111 (46%)
Frame = +1
Query: 303 MQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFN 362
M+ Y P +Q+ +F+ + VR + H + + +A +WF + PF+
Sbjct: 40 MEGLYLAGLPLVQQYLFQFECSVREHLPKLGEHFSYEMINPSMYASQWFITVFSYSFPFH 219
Query: 363 MVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLP 413
+ R+WD +L EG + + + + L D L KL F+ L+ L++ P
Sbjct: 220 LALRIWDVFLYEGVKI---VFKVGLALLKYCHDDLIKLPFEKLIHALKNFP 363
>TC232919
Length = 457
Score = 44.3 bits (103), Expect = 1e-04
Identities = 31/99 (31%), Positives = 49/99 (49%), Gaps = 7/99 (7%)
Frame = +2
Query: 112 SSMGARATDSAR----VMKFTKVLSGTVVILDKLRELAWS-GVPDY--MRPTVWRLLLGY 164
SS A A D +R ++ LS V+ + +LR + S G+ D +RPT+W+LLLGY
Sbjct: 149 SSPTATADDISRHALFFVQLLAELSRKVIDMRELRRVVASQGISDAGALRPTLWKLLLGY 328
Query: 165 APPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSEDEI 203
PP+ L +KR +Y + P E+++
Sbjct: 329 LPPDRSLWSSELTKKRSQYKNFKDDLLTNPSVRNHEEDV 445
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,716,814
Number of Sequences: 63676
Number of extensions: 261064
Number of successful extensions: 1468
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1327
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1441
length of query: 445
length of database: 12,639,632
effective HSP length: 100
effective length of query: 345
effective length of database: 6,272,032
effective search space: 2163851040
effective search space used: 2163851040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0106.13


