

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0104.2
(456 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
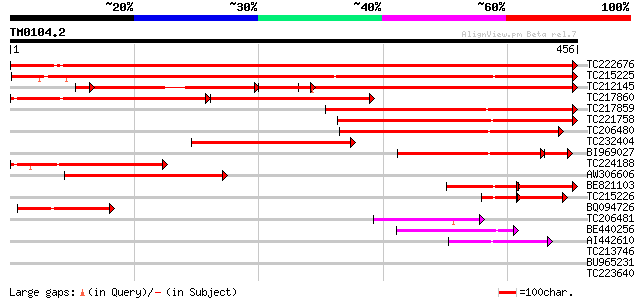
Score E
Sequences producing significant alignments: (bits) Value
TC222676 UP|Q84L88 (Q84L88) Apyrase, complete 641 0.0
TC215225 UP|Q9FVC3 (Q9FVC3) Apyrase GS50 (Fragment), complete 544 e-155
TC212145 UP|Q76IB9 (Q76IB9) Apyrase (Fragment), complete 298 e-127
TC217860 similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (56%) 209 e-107
TC217859 weakly similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, part... 275 2e-74
TC221758 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (... 258 4e-69
TC206480 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, p... 217 1e-56
TC232404 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, par... 181 5e-46
BI969027 homologue to GP|11225137|gb apyrase GS52 {Glycine soja}... 148 5e-42
TC224188 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (... 157 9e-39
AW306606 similar to GP|11225135|gb apyrase GS50 {Glycine soja}, ... 145 3e-35
BE821103 GP|11225135|gb apyrase GS50 {Glycine soja}, partial (23%) 82 9e-34
TC215226 similar to UP|Q9SPM7 (Q9SPM7) Apyrase, partial (15%) 57 6e-15
BQ094726 similar to GP|19352177|db PsAPY2 {Pisum sativum}, parti... 78 9e-15
TC206481 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, par... 74 1e-13
BE440256 weakly similar to GP|27804684|gb| apyrase-like protein ... 68 7e-12
AI442610 53 2e-07
TC213746 similar to UP|Q9LVF3 (Q9LVF3) Gb|AAD13707.1, partial (35%) 33 0.20
BU965231 30 1.7
TC223640 similar to UP|Q9FNL4 (Q9FNL4) Serine/threonine protein ... 30 3.0
>TC222676 UP|Q84L88 (Q84L88) Apyrase, complete
Length = 1368
Score = 641 bits (1654), Expect = 0.0
Identities = 313/457 (68%), Positives = 378/457 (82%), Gaps = 1/457 (0%)
Frame = +1
Query: 1 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 60
M+ LI L+TF+ MPAI+SSQYLGNN+L +RKI L K+ E ++SYAV+FDAGSTGSR+H
Sbjct: 1 MELLIKLITFLLFSMPAITSSQYLGNNLLTSRKIFL-KHVE-ISSYAVVFDAGSTGSRIH 174
Query: 61 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 120
VY+F+QNLDLL + +E+Y+ + PGLSSYA NPE+AA+SLIPLL++AE+VVP QP T
Sbjct: 175 VYHFNQNLDLLHIGKGVEYYNKITPGLSSYANNPEQAAKSLIPLLEQAEDVVPDDLQPKT 354
Query: 121 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY 180
PV+LGATAGLRLL G+A+E ILQ+VRDMLSNRS NVQ DAVSI+DGTQEGSYLWVT+NY
Sbjct: 355 PVRLGATAGLRLLNGDASEKILQSVRDMLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNY 534
Query: 181 LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL 240
LG LGK++TKTVGV+DLGGGSVQM YAVS+ TAKNAPKV +G+DPYIKK+VL+G YDL
Sbjct: 535 ALGNLGKKYTKTVGVIDLGGGSVQMAYAVSKKTAKNAPKVADGDDPYIKKVVLKGIPYDL 714
Query: 241 YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK 300
YVHSYL +GREA RAEI K+ S NPC+LAGF+G YTYSG E+K +A SG+N N+C+
Sbjct: 715 YVHSYLHFGREASRAEILKLTPRSPNPCLLAGFNGIYTYSGEEFKATAYTSGANFNKCKN 894
Query: 301 IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVNKPNAKIR 359
KALK+N PCPYQNCTFGGIWNGGGG+GQKN F +SSF+YL ED G + + PN +R
Sbjct: 895 TIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKNPFASSSFFYLPEDTGMVDASTPNFILR 1074
Query: 360 PVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTV 419
PVD++T AK AC N EDAKS YP L +K+ YVC+DL+Y Y LLVDGFGLDP Q++T
Sbjct: 1075PVDIETKAKEACALNFEDAKSTYPFLDKKNVASYVCMDLIYQYVLLVDGFGLDPLQKITS 1254
Query: 420 ANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
EIEYQDA+VEAAWPLG A+EAIS+LPKFERLMYF+
Sbjct: 1255GKEIEYQDAIVEAAWPLGNAVEAISALPKFERLMYFV 1365
>TC215225 UP|Q9FVC3 (Q9FVC3) Apyrase GS50 (Fragment), complete
Length = 1786
Score = 544 bits (1402), Expect = e-155
Identities = 280/464 (60%), Positives = 343/464 (73%), Gaps = 9/464 (1%)
Frame = +3
Query: 2 DFLISLMTFVFMLMPAISSSQ-----YLGNNILMNRKILLPKNQEPVT---SYAVIFDAG 53
D+LI L + ++ SSS YLG + + L P T SYAVIFDAG
Sbjct: 30 DYLIILSLLLSTVLATASSSSSSSSLYLGKGF--SHRKLSPSYHIHKTIDESYAVIFDAG 203
Query: 54 STGSRVHVYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVP 113
STGSRVHVY F+Q LDLL + +LE + PGLS+YA NP++AAESLIPLL+EAE VP
Sbjct: 204 STGSRVHVYRFNQQLDLLRIGQDLELFVKTMPGLSAYAENPQDAAESLIPLLEEAEAAVP 383
Query: 114 VSQQPNTPVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSY 173
P TPVKLGATAGLR LEG+A++ ILQAV DML NRS LNV +DAVS+L G QEG+Y
Sbjct: 384 QEFHPRTPVKLGATAGLRQLEGDASDRILQAVSDMLKNRSTLNVGADAVSVLSGNQEGAY 563
Query: 174 LWVTINYLLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVL 233
WVTINYLLG LGK +++TV VVDLGGGSVQM YAVS A AP+ P+G + YI ++ L
Sbjct: 564 QWVTINYLLGNLGKHYSETVAVVDLGGGSVQMAYAVSETDAAKAPRAPDGVESYITEMFL 743
Query: 234 QGKKYDLYVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGS 293
+GKKY LYVHSYLRYG A RAE KV S NPCILAGFDG Y Y G +YK AP SGS
Sbjct: 744 RGKKYYLYVHSYLRYGMLAARAEALKVR-DSENPCILAGFDGYYVYGGVQYKAKAPPSGS 920
Query: 294 NLNQCRKIALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-IFVN 352
+ +QC+ + ++AL VNA C Y++CTFGGIWNGGGG+G+ N F+ S F+ ++++ G + N
Sbjct: 921 SFSQCQNVVVEALHVNATCSYKDCTFGGIWNGGGGAGENNFFIASFFFEVADEAGFVDPN 1100
Query: 353 KPNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLD 412
PNAK+RPVD + AAK+AC T L+D KS +P + + D V Y+CLDLVY YTLLVDGFG+D
Sbjct: 1101APNAKVRPVDFENAAKVACNTELKDLKSIFPRVKDGD-VPYICLDLVYEYTLLVDGFGID 1277
Query: 413 PFQEVTVANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
P QE+T+ ++EYQD+LVEAAWPLG+AIEAISSLPKFE+LMYFI
Sbjct: 1278PQQEITLVRQVEYQDSLVEAAWPLGSAIEAISSLPKFEKLMYFI 1409
>TC212145 UP|Q76IB9 (Q76IB9) Apyrase (Fragment), complete
Length = 1173
Score = 298 bits (764), Expect(3) = e-127
Identities = 142/227 (62%), Positives = 175/227 (76%), Gaps = 3/227 (1%)
Frame = +1
Query: 233 LQGKKYDLYV--HSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPA 290
L ++Y + V +SYL +GREA RAEI K+ S NPC+LAG +G YTYSG E+K +A
Sbjct: 490 LYSREYHMIVMFNSYLHFGREASRAEILKLTPRSPNPCLLAGLNGIYTYSGEEFKATAYT 669
Query: 291 SGSNLNQCRKIALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVG-I 349
SG+N N+C+ KALK+N PCPYQNCTFGGIWNGGGG+GQKNLF +SSF+YL ED G +
Sbjct: 670 SGANFNKCKNTIRKALKLNYPCPYQNCTFGGIWNGGGGNGQKNLFASSSFFYLPEDTGMV 849
Query: 350 FVNKPNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGF 409
+ PN +RPVD++T AK AC N EDAKS YP L +K+ YVC+DL+Y Y LLVDGF
Sbjct: 850 DASTPNFILRPVDIETKAKKACALNFEDAKSTYPFLDKKNVASYVCMDLIYQYVLLVDGF 1029
Query: 410 GLDPFQEVTVANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
GLDP Q++T EIEYQ+A+VEAAWPLG A+EAIS+LPKFERLMYF+
Sbjct: 1030GLDPLQKITSGKEIEYQEAIVEAAWPLGNAVEAISALPKFERLMYFV 1170
Score = 165 bits (417), Expect(3) = e-127
Identities = 84/136 (61%), Positives = 102/136 (74%)
Frame = +2
Query: 66 QNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLG 125
+ LDLL + +E+Y+ + PGLSSYA NPE+AA+SLIPLL++AE+VVP QP TPV+LG
Sbjct: 38 RTLDLLHIGKGVEYYNKITPGLSSYANNPEQAAKSLIPLLEQAEDVVPDDLQPKTPVRLG 217
Query: 126 ATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKL 185
ILQ+ RDMLSNRS NVQ DAVSI+DGTQEGSYLWVT+NY G L
Sbjct: 218 ---------------ILQSGRDMLSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYAWGNL 352
Query: 186 GKRFTKTVGVVDLGGG 201
GK++TKTVGV+DLGGG
Sbjct: 353 GKKYTKTVGVIDLGGG 400
Score = 64.3 bits (155), Expect = 1e-10
Identities = 31/46 (67%), Positives = 36/46 (77%)
Frame = +3
Query: 201 GSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHSYL 246
GSVQM AVS+ TAKNAPKV +G+DPYIKK+VL+G YD YV L
Sbjct: 399 GSVQMADAVSKKTAKNAPKVADGDDPYIKKVVLKGIPYDRYVQQLL 536
Score = 31.2 bits (69), Expect(3) = e-127
Identities = 12/15 (80%), Positives = 15/15 (100%)
Frame = +1
Query: 54 STGSRVHVYNFDQNL 68
STGSR+HVY+F+QNL
Sbjct: 1 STGSRIHVYHFNQNL 45
>TC217860 similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (56%)
Length = 881
Score = 209 bits (532), Expect(2) = e-107
Identities = 108/161 (67%), Positives = 134/161 (83%)
Frame = +3
Query: 1 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 60
M+FL + +T + ++PA SSSQYLGNN+L +RKI L K+ +TSYAVIFD GSTG+RVH
Sbjct: 6 MNFL-TFVTVLLFILPATSSSQYLGNNLLTHRKIFLKKDNI-ITSYAVIFDGGSTGTRVH 179
Query: 61 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 120
V++FDQNLDLLP+ + LE + PGLS+Y +PE+AAESLIPLL+EAE+VVP +PNT
Sbjct: 180 VFHFDQNLDLLPIGDSLELNKKITPGLSAYEDDPEQAAESLIPLLEEAESVVPEDLRPNT 359
Query: 121 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDA 161
PV+LGATAGLRLL+GNA+E ILQAVRDMLSNRS LN+QSDA
Sbjct: 360 PVRLGATAGLRLLKGNASEQILQAVRDMLSNRSTLNLQSDA 482
Score = 198 bits (503), Expect(2) = e-107
Identities = 94/132 (71%), Positives = 108/132 (81%)
Frame = +1
Query: 162 VSILDGTQEGSYLWVTINYLLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVP 221
V+ILDG QE +Y+WV +NYLLG LGK +KTVGV DLGGGSVQM YAVS+NTAKNAP+ P
Sbjct: 484 VTILDGNQEAAYMWVALNYLLGNLGKVISKTVGVADLGGGSVQMAYAVSKNTAKNAPQPP 663
Query: 222 EGEDPYIKKLVLQGKKYDLYVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSG 281
EGE+ YIK LVL GK YDLYVHSYL +G+EA RAE+ KV G SANPCILAG++G YTYSG
Sbjct: 664 EGEESYIKTLVLNGKTYDLYVHSYLHFGKEASRAEMLKVTGDSANPCILAGYNGTYTYSG 843
Query: 282 AEYKVSAPASGS 293
+YK A SGS
Sbjct: 844 VKYKALASTSGS 879
>TC217859 weakly similar to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (41%)
Length = 685
Score = 275 bits (704), Expect = 2e-74
Identities = 131/203 (64%), Positives = 160/203 (78%), Gaps = 1/203 (0%)
Frame = +2
Query: 255 AEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALKALKVNAPCPY 314
AE+ V G S NPCI+A +DG YTYSG EYK A GSN ++CR++ALKALKVN PCP+
Sbjct: 2 AEMSNVTGDSPNPCIVAAYDGTYTYSGVEYKALASTCGSNFDKCREVALKALKVNEPCPH 181
Query: 315 QNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIF-VNKPNAKIRPVDLKTAAKLACKT 373
QNCTFGGIWNGGGGSGQK L++T+SFYYL GI +K ++K+ P + K AAK AC+
Sbjct: 182 QNCTFGGIWNGGGGSGQKVLYVTTSFYYLVIQAGIADASKTSSKVYPAEFKAAAKRACQV 361
Query: 374 NLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANEIEYQDALVEAA 433
EDA+S YP L +D++ Y+C+D+ Y YTLLVDGFGLDP++E+ VANEIEYQ ALVE A
Sbjct: 362 KFEDAQSTYP-LMMEDALPYICMDITYQYTLLVDGFGLDPWKEIIVANEIEYQGALVEGA 538
Query: 434 WPLGTAIEAISSLPKFERLMYFI 456
WPLG+AIEAISSLPKF +LMYFI
Sbjct: 539 WPLGSAIEAISSLPKFNKLMYFI 607
>TC221758 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (37%)
Length = 625
Score = 258 bits (659), Expect = 4e-69
Identities = 129/194 (66%), Positives = 152/194 (77%), Gaps = 1/194 (0%)
Frame = +1
Query: 264 SANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALKALKVNAPCPYQNCTFGGIW 323
SA CI AGFDG YTY ++S P ++CR++ L+ALK+N CP+Q CTFGGIW
Sbjct: 1 SATLCI*AGFDGDYTYQREIIRLSLPFLAPVKDECREVVLQALKLNKSCPHQYCTFGGIW 180
Query: 324 NGGGGSGQKNLFLTSSFYYLSEDVGIF-VNKPNAKIRPVDLKTAAKLACKTNLEDAKSKY 382
+GG GSGQK LF TSSFYYL ++GI +NKPN+KI PVDL+ AK AC+T LEDAKS Y
Sbjct: 181 DGGRGSGQKILFGTSSFYYLPTEIGIIDLNKPNSKIHPVDLEIEAKRACETKLEDAKSTY 360
Query: 383 PDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANEIEYQDALVEAAWPLGTAIEA 442
P+L E D + YVCLD+ Y Y L DGFGLDP+QE+TVANEIEYQDALVEAAWPLGTAIEA
Sbjct: 361 PNLAE-DRLPYVCLDIAYQYALYTDGFGLDPWQEITVANEIEYQDALVEAAWPLGTAIEA 537
Query: 443 ISSLPKFERLMYFI 456
ISSLPKF+RLM+FI
Sbjct: 538 ISSLPKFDRLMFFI 579
>TC206480 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (39%)
Length = 906
Score = 217 bits (552), Expect = 1e-56
Identities = 100/182 (54%), Positives = 141/182 (76%), Gaps = 2/182 (1%)
Frame = +1
Query: 266 NPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALKALKVN-APCPYQNCTFGGIWN 324
NPCI++G+DG+Y Y G +K S+ +SG++LN+C+ +AL+ALKVN + C + CTFGGIWN
Sbjct: 13 NPCIISGYDGSYNYGGKSFKASSGSSGASLNECKSVALRALKVNESTCTHMKCTFGGIWN 192
Query: 325 GGGGSGQKNLFLTSSFYYLSEDVGIF-VNKPNAKIRPVDLKTAAKLACKTNLEDAKSKYP 383
GGGG GQKNLF+ S F+ + + G N P A +RP D + AAK AC+T LE+AKS +P
Sbjct: 193 GGGGDGQKNLFVASFFFDRAAEAGFADPNLPVAIVRPADFEDAAKQACQTKLENAKSTFP 372
Query: 384 DLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANEIEYQDALVEAAWPLGTAIEAI 443
+ ++ ++ Y+C+DL+Y YTLLVDGFG+ P+QE+T+ +++Y DALVEAAWPLG+AIEA+
Sbjct: 373 RV-DEGNLPYLCMDLIYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAV 549
Query: 444 SS 445
SS
Sbjct: 550 SS 555
>TC232404 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (28%)
Length = 535
Score = 181 bits (460), Expect = 5e-46
Identities = 84/132 (63%), Positives = 107/132 (80%)
Frame = +1
Query: 147 DMLSNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKLGKRFTKTVGVVDLGGGSVQMT 206
D+L +RS+L +SDAV++LDGTQEG+Y WVTINYLLG LGK ++KTVGVVDLGGGSVQM
Sbjct: 1 DLLKDRSSLKSESDAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMA 180
Query: 207 YAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDLYVHSYLRYGREAFRAEIFKVAGGSAN 266
YA+S A APKV +G DPY+K++ L+G+KY LYVHSYL YG A RAEI K++ + N
Sbjct: 181 YAISETDAAMAPKVVDGGDPYLKEMFLRGRKYYLYVHSYLHYGLLAARAEILKISDDAEN 360
Query: 267 PCILAGFDGAYT 278
PC+++G+DG YT
Sbjct: 361 PCVISGYDGKYT 396
>BI969027 homologue to GP|11225137|gb apyrase GS52 {Glycine soja}, partial
(29%)
Length = 423
Score = 148 bits (374), Expect(2) = 5e-42
Identities = 76/121 (62%), Positives = 90/121 (73%), Gaps = 2/121 (1%)
Frame = -1
Query: 313 PYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIF-VNKPNAKIRPVDLKTAAKLAC 371
P+QNC FGGIW+GG GSGQK LF TSSFYYL +GI +NKPN KI PVDL+ K AC
Sbjct: 423 PHQNCAFGGIWDGGRGSGQKILFGTSSFYYLPTXIGIIDLNKPNCKIHPVDLEIEVKRAC 244
Query: 372 KTNLEDAKSKYPDLYEKDSVEYVCLDLVYVY-TLLVDGFGLDPFQEVTVANEIEYQDALV 430
+ LE AKS YP+L ++D + YVCLD+ DGFGLDP+QE+TVANEIEYQDALV
Sbjct: 243 GSKLEXAKSTYPNL-DEDRLPYVCLDIAXXXCXCTXDGFGLDPWQEITVANEIEYQDALV 67
Query: 431 E 431
+
Sbjct: 66 K 64
Score = 41.2 bits (95), Expect(2) = 5e-42
Identities = 19/22 (86%), Positives = 21/22 (95%)
Frame = -3
Query: 431 EAAWPLGTAIEAISSLPKFERL 452
EAAWPLGTAIEAISSL KF+R+
Sbjct: 67 EAAWPLGTAIEAISSLLKFDRV 2
>TC224188 homologue to UP|Q9FVC2 (Q9FVC2) Apyrase GS52, partial (28%)
Length = 407
Score = 157 bits (397), Expect = 9e-39
Identities = 84/129 (65%), Positives = 101/129 (78%), Gaps = 2/129 (1%)
Frame = +3
Query: 1 MDFLISLMTFVFMLM--PAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSR 58
MDFL +L T + +L A+SS+QY NIL+ + + PK QE +TSYAVIFDAGSTGSR
Sbjct: 27 MDFL-TLFTLLLLLFIHTALSSTQYHDGNILLTHRKIFPK-QEAITSYAVIFDAGSTGSR 200
Query: 59 VHVYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQP 118
VHV++FDQNLDLL + NELEFYD V PGLS+Y NP++AAESLIPLL+EAE+VVP P
Sbjct: 201 VHVFHFDQNLDLLHIGNELEFYDKVTPGLSAYTDNPQQAAESLIPLLEEAESVVPEDLYP 380
Query: 119 NTPVKLGAT 127
TPVKLGAT
Sbjct: 381 TTPVKLGAT 407
>AW306606 similar to GP|11225135|gb apyrase GS50 {Glycine soja}, partial
(33%)
Length = 467
Score = 145 bits (367), Expect = 3e-35
Identities = 78/131 (59%), Positives = 94/131 (71%)
Frame = +3
Query: 45 SYAVIFDAGSTGSRVHVYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPL 104
SYAVIFDAGSTGSRVHVY F+Q LDLL + LE PGLS+YA NP++AA SLI L
Sbjct: 75 SYAVIFDAGSTGSRVHVYRFNQQLDLLLIGQYLELLVKTMPGLSAYAENPQDAA*SLISL 254
Query: 105 LKEAENVVPVSQQPNTPVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSI 164
L+EA VP P TPVKL ATA L+ L+ +A++ ILQAV DML NRS LNV++DA ++
Sbjct: 255 LEEA*AAVPQELHPMTPVKLVATARLKHLKSDASDKILQAVNDMLKNRSTLNVEADADAV 434
Query: 165 LDGTQEGSYLW 175
L QE +Y W
Sbjct: 435 LSANQERAYQW 467
>BE821103 GP|11225135|gb apyrase GS50 {Glycine soja}, partial (23%)
Length = 501
Score = 81.6 bits (200), Expect(2) = 9e-34
Identities = 36/47 (76%), Positives = 44/47 (93%)
Frame = -3
Query: 410 GLDPFQEVTVANEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
G+DP QE+T+ ++EYQD+LVEAAWPLG+AIEAISSLPKFE+LMYFI
Sbjct: 193 GIDPQQEITLVRQVEYQDSLVEAAWPLGSAIEAISSLPKFEKLMYFI 53
Score = 80.5 bits (197), Expect(2) = 9e-34
Identities = 38/60 (63%), Positives = 47/60 (78%)
Frame = -2
Query: 352 NKPNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGL 411
N PNAK+RPVD + AAK+AC T L+D KS +P + + D V Y+CLDLVY YTLLVDGFG+
Sbjct: 491 NAPNAKVRPVDFENAAKVACNTELKDLKSIFPRVKDGD-VPYICLDLVYEYTLLVDGFGM 315
>TC215226 similar to UP|Q9SPM7 (Q9SPM7) Apyrase, partial (15%)
Length = 331
Score = 56.6 bits (135), Expect(2) = 6e-15
Identities = 26/40 (65%), Positives = 33/40 (82%)
Frame = +2
Query: 409 FGLDPFQEVTVANEIEYQDALVEAAWPLGTAIEAISSLPK 448
F + QE+T+ ++EY D+LVEAAWPLG+AIEAISSLPK
Sbjct: 212 FRIKSQQEITLVKQVEY*DSLVEAAWPLGSAIEAISSLPK 331
Score = 42.0 bits (97), Expect(2) = 6e-15
Identities = 20/32 (62%), Positives = 25/32 (77%)
Frame = +1
Query: 380 SKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGL 411
S +P + + D V Y+CLDLVY YTLLVDGFG+
Sbjct: 1 SIFPRVKDGD-VPYICLDLVYEYTLLVDGFGM 93
>BQ094726 similar to GP|19352177|db PsAPY2 {Pisum sativum}, partial (22%)
Length = 421
Score = 77.8 bits (190), Expect = 9e-15
Identities = 39/78 (50%), Positives = 58/78 (74%)
Frame = +1
Query: 7 LMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVHVYNFDQ 66
L+TFV +MP+ +S++ +G+ L+NRK + ++ V SYAVIFDAGS+GSRVHV++FD
Sbjct: 127 LITFVLYVMPSSTSNESVGDYALVNRK--MSPEKKSVGSYAVIFDAGSSGSRVHVFHFDH 300
Query: 67 NLDLLPVENELEFYDSVK 84
NLDL+ + +LE + VK
Sbjct: 301 NLDLVHIGKDLELFVQVK 354
>TC206481 similar to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (11%)
Length = 315
Score = 74.3 bits (181), Expect = 1e-13
Identities = 41/93 (44%), Positives = 54/93 (57%), Gaps = 3/93 (3%)
Frame = +2
Query: 293 SNLNQCRKIALKALKVN-APCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFV 351
++LN+C+ IA +ALKVN + C + CTFGGIWNGGGG GQKNLF+ S F+ +
Sbjct: 2 ASLNECKNIAFQALKVNESKCTHMKCTFGGIWNGGGGDGQKNLFVDSFFFDRGR*CWFCL 181
Query: 352 NKPN--AKIRPVDLKTAAKLACKTNLEDAKSKY 382
+K + P + ACKT L A S Y
Sbjct: 182 SKFTRCEGLVPWIFRLQLSKACKTKLAYATSTY 280
>BE440256 weakly similar to GP|27804684|gb| apyrase-like protein {Medicago
truncatula}, partial (20%)
Length = 298
Score = 68.2 bits (165), Expect = 7e-12
Identities = 39/99 (39%), Positives = 55/99 (55%), Gaps = 1/99 (1%)
Frame = +2
Query: 312 CPYQNCTFGGIWNGGGGSGQKNLFLTSSFY-YLSEDVGIFVNKPNAKIRPVDLKTAAKLA 370
C + CTF GIWNGGGG G KNLF+ S F+ SE N P P ++ A
Sbjct: 5 CTHMKCTFRGIWNGGGGDGHKNLFVASLFFDRSSEACSTDPNLPVVIYHPANIYDTLTHA 184
Query: 371 CKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGF 409
C+T LE+A P +++ + + Y +DL+Y +TLL D +
Sbjct: 185 CQT*LENA*FTLPLVHDLNLI-YHRIDLIYHHTLLQDPY 298
>AI442610
Length = 253
Score = 53.1 bits (126), Expect = 2e-07
Identities = 28/83 (33%), Positives = 46/83 (54%)
Frame = +2
Query: 354 PNAKIRPVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDP 413
P +R + A AC + L+ AK +P + + + Y+ ++L+Y +TLL DGFGL
Sbjct: 8 PALIVRTAHFEDATNPACLSKLDSAKYIFPRVAD-GYLNYISMELIYQHTLLADGFGLYT 184
Query: 414 FQEVTVANEIEYQDALVEAAWPL 436
++ +T + + D L EAAW L
Sbjct: 185 WKYITSVIKSKNDDVLFEAAWSL 253
>TC213746 similar to UP|Q9LVF3 (Q9LVF3) Gb|AAD13707.1, partial (35%)
Length = 959
Score = 33.5 bits (75), Expect = 0.20
Identities = 13/28 (46%), Positives = 19/28 (67%)
Frame = +1
Query: 285 KVSAPASGSNLNQCRKIALKALKVNAPC 312
KV+APA + + QCRK+ K +K+ PC
Sbjct: 490 KVNAPAQKNVIRQCRKLKRKVVKITPPC 573
>BU965231
Length = 445
Score = 30.4 bits (67), Expect = 1.7
Identities = 11/19 (57%), Positives = 15/19 (78%)
Frame = +1
Query: 46 YAVIFDAGSTGSRVHVYNF 64
Y +I D GSTG+RVHV+ +
Sbjct: 331 YRIIVDGGSTGTRVHVFKY 387
>TC223640 similar to UP|Q9FNL4 (Q9FNL4) Serine/threonine protein kinase-like
protein, partial (16%)
Length = 645
Score = 29.6 bits (65), Expect = 3.0
Identities = 16/38 (42%), Positives = 21/38 (55%)
Frame = -1
Query: 5 ISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEP 42
ISLMT+ L P +SSS +++L R L PK P
Sbjct: 333 ISLMTWS*SLFPCLSSSSPSSSSLLARRSPLNPKKHSP 220
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,568,912
Number of Sequences: 63676
Number of extensions: 220246
Number of successful extensions: 1234
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1201
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1210
length of query: 456
length of database: 12,639,632
effective HSP length: 100
effective length of query: 356
effective length of database: 6,272,032
effective search space: 2232843392
effective search space used: 2232843392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0104.2


