

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.9
(705 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
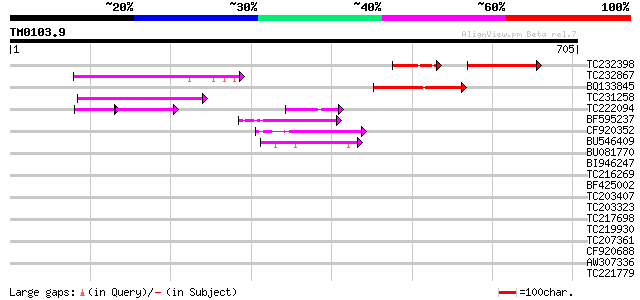
Score E
Sequences producing significant alignments: (bits) Value
TC232398 94 2e-28
TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial ... 110 2e-24
BQ133845 102 4e-22
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 94 3e-19
TC222094 51 5e-13
BF595237 68 1e-11
CF920352 67 2e-11
BU546409 62 1e-09
BU081770 40 0.005
BI946247 34 0.19
TC216269 similar to UP|E6_GOSHI (Q01197) Protein E6, partial (17%) 33 0.56
BF425002 32 0.96
TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pr... 30 2.8
TC203323 weakly similar to UP|Q7QHT5 (Q7QHT5) AgCP6936, partial ... 30 2.8
TC217698 similar to UP|Q89GY6 (Q89GY6) Blr6209 protein, partial ... 30 3.7
TC219930 weakly similar to GB|AAP37844.1|30725644|BT008485 At3g1... 30 3.7
TC207361 similar to UP|Q9FMU9 (Q9FMU9) Similarity to ATFP3, part... 30 3.7
CF920688 30 4.8
AW307336 weakly similar to GP|22202765|dbj P0039H02.8 {Oryza sat... 30 4.8
TC221779 30 4.8
>TC232398
Length = 1054
Score = 93.6 bits (231), Expect(2) = 2e-28
Identities = 44/92 (47%), Positives = 67/92 (72%)
Frame = +1
Query: 570 RLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILG 629
R+ +++PT M LQLAD SI +GVVED+LV+V + F VDFVI+D++ED++I LILG
Sbjct: 175 RIGNQKIEPTRMTLQLADHSITRSFGVVEDILVKVHQLIFLVDFVIMDIEEDAEIRLILG 354
Query: 630 RPFLATSQAKINVGNGTISLRVADEKITFTIF 661
PF+ T++ +++G G + + V D+K TF +F
Sbjct: 355 WPFMVTAKCVVDMGKGNLEMSVEDQKATFNLF 450
Score = 51.6 bits (122), Expect(2) = 2e-28
Identities = 28/60 (46%), Positives = 40/60 (66%)
Frame = +2
Query: 477 LRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPG 536
L + +PF E +++MP Y KF+K+IL KK + +E I + E C A++Q KLPPK KD G
Sbjct: 2 LEITIPFGERIQQMPLYKKFLKDILIKKGKYI-NSETIVVGEYCRALIQ-KLPPKFKDLG 175
>TC232867 similar to UP|Q9MA68 (Q9MA68) F2J6.13 protein, partial (3%)
Length = 1144
Score = 110 bits (275), Expect = 2e-24
Identities = 72/228 (31%), Positives = 116/228 (50%), Gaps = 16/228 (7%)
Frame = -3
Query: 80 AELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFK 139
A+ +RL L FSL A+ WL+S S+ +W+++ +KF KK FP + + K I F
Sbjct: 695 ADAIRLSLLSFSLSGEAKRWLHSFKGNSLKSWDEVVEKFLKKYFPESKTAEGKAAISSFH 516
Query: 140 QEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSA 199
Q E+L EALERF+ LLRK P H + +Q+ F D L ++ ++A+ G+ + +
Sbjct: 515 QFPDESLSEALERFRGLLRKTPTHGFSEPIQLNIFIDELRPESKQLVDASVGGKIKMKTP 336
Query: 200 QAGWNLINKMAESAVNSTNDRQN---RRGVLEIEAYDRMVASNKQLSQQMTTIQR----- 251
+LI MA S + DR + ++ +LE+ + D ++A NK LS+Q+ T+ +
Sbjct: 335 DEAMDLIESMAASDIAILRDRAHIPTKKSLLELTSQDTLLAQNKLLSKQLETLTKTLSKL 156
Query: 252 --QFQAAKISNVDSIH---CGTCGGPHASEEC---GTYFDEEVKVLGN 291
Q +A+ S+ + C G H S C EV +GN
Sbjct: 155 PTQLHSAQTSHSSILQVTGCTIFGEAHESGCCIPNEEQIAHEVNYMGN 12
>BQ133845
Length = 389
Score = 102 bits (255), Expect = 4e-22
Identities = 51/115 (44%), Positives = 76/115 (65%)
Frame = +2
Query: 453 PFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENE 512
P+P + + E+ +KF+ +FKKL + LPF E L++MP YA F+K++L+KK ++
Sbjct: 38 PYPLVPS*KDKEQHLAKFLDIFKKLEITLPFEEALQQMPLYANFLKDMLTKKNWYIHSDK 217
Query: 513 IIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSM 567
I+ + CSA++QR LPP DPG T+P + G +AL DLG+S+NLMPLSM
Sbjct: 218 IV-VEGNCSAVIQRILPP*HTDPGFVTMPCSIGEVAVGKALIDLGASINLMPLSM 379
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 93.6 bits (231), Expect = 3e-19
Identities = 54/164 (32%), Positives = 85/164 (50%), Gaps = 2/164 (1%)
Frame = +2
Query: 85 LQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDTE 144
L+ FP SL A++WL SI W+DL + F +K FP + ++ DI +Q E
Sbjct: 656 LKAFPHSLEGVAKDWLYYLAPRSIFNWDDLKRVFLEKFFPASRTTTIRKDISGIRQLSRE 835
Query: 145 NLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGWN 204
+L+E ERFKK CP H ++ + + FY+ L++ RS ++AA+ G ++ N
Sbjct: 836 SLYEYWERFKKSCASCPHHQISKQLLL*YFYEELSNMKRSMIDAASGGALGDMTPTEARN 1015
Query: 205 LINKMAESA--VNSTNDRQNRRGVLEIEAYDRMVASNKQLSQQM 246
LI KMA ++ ++ ND RGV E+ NK L + +
Sbjct: 1016LIKKMASNSQQFSARNDAIVLRGVHEVATDSSSSTENKSLRENL 1147
>TC222094
Length = 984
Score = 50.8 bits (120), Expect(2) = 5e-13
Identities = 29/79 (36%), Positives = 43/79 (53%)
Frame = -3
Query: 132 KNDILVFKQEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAAS 191
K +I F Q E+L EAL+RF LL K P H + VQ+ F DG+ ++ L+A+A
Sbjct: 478 KVEISSFHQHPHESLSEALDRFHGLLWKTPTHGFSEPVQLNIFIDGMQPHSKQLLDASAG 299
Query: 192 GEFEALSAQAGWNLINKMA 210
G+ + + + LI MA
Sbjct: 298 GKIKLKTPEEAIELIENMA 242
Score = 42.7 bits (99), Expect = 5e-04
Identities = 24/73 (32%), Positives = 40/73 (53%)
Frame = -3
Query: 343 QNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQVGQMAKQMSERPPGMFPS 402
++Q+ LEEL+ F+ + + K+ +AAI++LE Q+AK + ERP F
Sbjct: 220 RDQEPSPQEESTRLEELLVQFMQETRSHQKSTDAAIRNLEV---QLAKLVPERPTETFAV 50
Query: 403 DTVINPKENCSAI 415
+T + PK+ C I
Sbjct: 49 NTKMKPKKECKVI 11
Score = 42.0 bits (97), Expect(2) = 5e-13
Identities = 23/58 (39%), Positives = 31/58 (52%), Gaps = 2/58 (3%)
Frame = -2
Query: 81 ELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFP--RTLLRKLKNDIL 136
+ + L LF FSL A WL S ++ TW + +KF KK FP +T+ K N IL
Sbjct: 632 DAIHLDLFCFSLAGEARTWLRSFKGNNLRTWNEXXEKFLKKYFPESKTIXXKGGNLIL 459
>BF595237
Length = 415
Score = 68.2 bits (165), Expect = 1e-11
Identities = 47/130 (36%), Positives = 74/130 (56%), Gaps = 2/130 (1%)
Frame = +3
Query: 285 EVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFNQGGNNQRQYSNQRFQSQNSGPQQN 344
EV +G +QN+ YN G P F+ + NF QG + + NQ + Q S P QN
Sbjct: 33 EVNYMG-AQNHHGFQGYNQG--GPPGFN--QGRNFTQG*SWKNHPGNQFNKEQRSQPIQN 197
Query: 345 QDQGSGNGKKS--LEELMENFINKADTSFKNHEAAIKSLETQVGQMAKQMSERPPGMFPS 402
+Q +K+ LEE + F+ +++ + +++IK+LE Q+GQ+AKQM+ERP F +
Sbjct: 198 SNQVVNLYEKTSKLEETLNQFM*MFMSNYMSTKSSIKNLEIQMGQLAKQMAERPTNNFGA 377
Query: 403 DTVINPKENC 412
+T NPK+ C
Sbjct: 378 NTEKNPKKEC 407
>CF920352
Length = 571
Score = 67.4 bits (163), Expect = 2e-11
Identities = 49/141 (34%), Positives = 77/141 (53%), Gaps = 3/141 (2%)
Frame = -2
Query: 306 RNHPNFSWREQNNFNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSG--NGKKSLEELMENF 363
RNHP Q N +QGG++ R PQQ QGS + LEE + F
Sbjct: 477 RNHPG----NQFNGDQGGSSIR-------------PQQ---QGSSLYDRTTKLEETLAQF 358
Query: 364 INKADTSFKNHEAAIKSLETQVGQMAKQMSERPPGMFPSDTVINPKENCSAITLRN-IEP 422
+ + ++ K+ E+AIK+LE QVGQ+AK++++R F ++T NPKE C A+ R+ +
Sbjct: 357 M*VSMSNQKSTESAIKNLEVQVGQLAKKLADRSSSSFSANTEKNPKEECKAVMTRSKMTT 178
Query: 423 LGENKKKQKEKEEEKRKVELE 443
+ K +K+ EE K+++ E
Sbjct: 177 HVDEGKAEKKMEEHKQQLATE 115
>BU546409
Length = 528
Score = 61.6 bits (148), Expect = 1e-09
Identities = 43/144 (29%), Positives = 71/144 (48%), Gaps = 18/144 (12%)
Frame = -1
Query: 313 WREQNNFNQGGNNQRQ---YSNQRFQSQNSGPQQNQDQGSGNGKK------------SLE 357
W + +QGG + Q Y+ Q + Q N+DQG + K LE
Sbjct: 495 WEINRDKDQGGFSGFQ*GPYNQQG*WRSHPSNQFNKDQGGPSNKPIQQGPNIFQRTXKLE 316
Query: 358 ELMENFINKADTSFKNHEAAIKSLETQVGQMAKQMSERPPGMFPSDTVINPKENCSAITL 417
E + F+ ++ K+ E+A+++LE QVGQ+AKQ++++ F +T NPKE C +
Sbjct: 315 ETLTQFMKVIMSNHKSTESALENLEVQVGQLAKQITDKSSNSFMVNTEKNPKEECKDVMT 136
Query: 418 RN---IEPLGENKKKQKEKEEEKR 438
R+ +E E+ K+K EK+
Sbjct: 135 RSKRFVEAEDEDNVVSKKKFAEKK 64
>BU081770
Length = 426
Score = 39.7 bits (91), Expect = 0.005
Identities = 32/97 (32%), Positives = 50/97 (50%), Gaps = 3/97 (3%)
Frame = +1
Query: 326 QRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQV 385
Q+Q Q+ Q+ + PQ SLEEL+ + + +A+I+SL Q+
Sbjct: 97 QQQQQPQK*QTVEAPPQP-----------SLEELVRQMTMQNIQFQQETKASIQSLTNQM 243
Query: 386 GQMAKQMSERP---PGMFPSDTVINPKENCSAITLRN 419
GQ+A Q++++ PS V NPK N SAI+LR+
Sbjct: 244 GQLATQLNQQQSQNSDKLPSQAVQNPK-NVSAISLRS 351
>BI946247
Length = 372
Score = 34.3 bits (77), Expect = 0.19
Identities = 14/31 (45%), Positives = 24/31 (77%)
Frame = +3
Query: 425 ENKKKQKEKEEEKRKVELENKFTKVLFPPFP 455
+ KKK+K+K+++K+K + + K T LFPP+P
Sbjct: 114 KKKKKKKKKKKKKKKKKKKKKKTXKLFPPWP 206
>TC216269 similar to UP|E6_GOSHI (Q01197) Protein E6, partial (17%)
Length = 1139
Score = 32.7 bits (73), Expect = 0.56
Identities = 19/70 (27%), Positives = 31/70 (44%), Gaps = 7/70 (10%)
Frame = +1
Query: 281 YFDEEVKVLGNSQNNPYSNTYNLGWRNHPN-------FSWREQNNFNQGGNNQRQYSNQR 333
Y +EE N+ NN Y N+Y + + N +S+ NN+N N Y+N
Sbjct: 445 YTEEEYN---NNNNNKYHNSYQNNNQKYYNNDAANGIYSYNNNNNYNANNNRYNTYNNNN 615
Query: 334 FQSQNSGPQQ 343
+ +G +Q
Sbjct: 616 AVNGYNGERQ 645
>BF425002
Length = 416
Score = 32.0 bits (71), Expect = 0.96
Identities = 26/94 (27%), Positives = 45/94 (47%), Gaps = 4/94 (4%)
Frame = +3
Query: 272 PHASEECGTYFDEEVKVLGNSQNNP---YSNTYNLGWRNHPNFSWREQNNFNQGGNNQRQ 328
P+ ++ C + E VK ++N P + N N+ + +HP+ ++F + +
Sbjct: 18 PYPTKPCHLWCCESVK----ARNQPS*CWHNPNNISYHSHPHPLMELSSSFLFSPLIRNR 185
Query: 329 YSNQRFQSQNSGPQQNQDQGSGNG-KKSLEELME 361
N R + PQQ QD SGN ++ L+EL E
Sbjct: 186 NPNFRIVAVALEPQQQQDFPSGNSPQRLLKELAE 287
>TC203407 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(AtAGP4) (AT5g10430/F12B17_220), partial (50%)
Length = 966
Score = 30.4 bits (67), Expect = 2.8
Identities = 15/36 (41%), Positives = 24/36 (66%), Gaps = 2/36 (5%)
Frame = -1
Query: 405 VINPKENCSAITLRNIEPLGENKK--KQKEKEEEKR 438
+INP ++ + N++ LGE +K K +E+EEEKR
Sbjct: 849 IINPPKSRMNKHINNVQKLGEKRKHIKPEEEEEEKR 742
>TC203323 weakly similar to UP|Q7QHT5 (Q7QHT5) AgCP6936, partial (21%)
Length = 1063
Score = 30.4 bits (67), Expect = 2.8
Identities = 15/48 (31%), Positives = 23/48 (47%)
Frame = -1
Query: 291 NSQNNPYSNTYNLGWRNHPNFSWREQNNFNQGGNNQRQYSNQRFQSQN 338
N+ N ++ N W N+ N W N+N+ N R Y RF ++N
Sbjct: 622 NNGNYLWNRLNNRNWLNNRN*FWFRLINWNRNRFNNRNYLWNRFNNRN 479
>TC217698 similar to UP|Q89GY6 (Q89GY6) Blr6209 protein, partial (5%)
Length = 865
Score = 30.0 bits (66), Expect = 3.7
Identities = 15/36 (41%), Positives = 16/36 (43%)
Frame = +2
Query: 316 QNNFNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGN 351
Q N+ Q N N SQN PQQN Q S N
Sbjct: 44 QXNYGQASQNYPPQQNYGMTSQNYTPQQNYGQASPN 151
>TC219930 weakly similar to GB|AAP37844.1|30725644|BT008485 At3g18210
{Arabidopsis thaliana;} , partial (46%)
Length = 700
Score = 30.0 bits (66), Expect = 3.7
Identities = 25/91 (27%), Positives = 40/91 (43%), Gaps = 9/91 (9%)
Frame = +1
Query: 472 SMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPK 531
++F L LP S + + A FM+EIL K L E N + E I+ P
Sbjct: 223 NIFSSLERYLPPSMLNVPRDEKANFMREILLKYLPLGERNRAQKHREYRERIIANYQPLH 402
Query: 532 RK----DPGSFTLP-----VNFGASKQVRAL 553
R+ DP +F +P +N + +R++
Sbjct: 403 RELYSVDPATFFVPTFLRAINDNTEQSIRSI 495
>TC207361 similar to UP|Q9FMU9 (Q9FMU9) Similarity to ATFP3, partial (53%)
Length = 1127
Score = 30.0 bits (66), Expect = 3.7
Identities = 14/36 (38%), Positives = 23/36 (63%)
Frame = +1
Query: 410 ENCSAITLRNIEPLGENKKKQKEKEEEKRKVELENK 445
E+ SA N P+G+ ++K +E + E++KVE E K
Sbjct: 1 EHQSAALNTNSTPMGQEEEKSQENKVEEKKVEEEEK 108
>CF920688
Length = 521
Score = 29.6 bits (65), Expect = 4.8
Identities = 26/77 (33%), Positives = 40/77 (51%), Gaps = 3/77 (3%)
Frame = +3
Query: 399 MFPSDTVINPKENCSAITL---RNIEPLGENKKKQKEKEEEKRKVELENKFTKVLFPPFP 455
+F DT + K +CSAI + R + KKK+K+K+++K+K + + K K FP
Sbjct: 72 LFVVDTFV--K*HCSAIMIVIFRT*XKKKKKKKKKKKKKKKKKKKKKKIKKKKKKKKKFP 245
Query: 456 TNIAKRRLEKQFSKFIS 472
A +R F KF S
Sbjct: 246 AGGAPQR--GNFGKFPS 290
>AW307336 weakly similar to GP|22202765|dbj P0039H02.8 {Oryza sativa
(japonica cultivar-group)}, partial (18%)
Length = 371
Score = 29.6 bits (65), Expect = 4.8
Identities = 25/94 (26%), Positives = 46/94 (48%), Gaps = 11/94 (11%)
Frame = +2
Query: 402 SDTVINPKENC---SAITLRNIEPLGENKKKQKEKEEEKRKVELENKFTKVLFPPF---- 454
SD +++ K N S++ + + + KKK+K+K+++KRK + + K +K++F
Sbjct: 20 SDVLLSLKVNACFLSSV*KKKKKKKKKKKKKKKKKKKKKRKKKKKKKTSKLVFFTIAKKG 199
Query: 455 --PTNIAKRRLEKQFSKFISMF--KKLRVELPFS 484
P + F F S KK + +PFS
Sbjct: 200 KQPKRVKWETKSPNFKNFFSRASQKKSK*IIPFS 301
>TC221779
Length = 475
Score = 29.6 bits (65), Expect = 4.8
Identities = 21/85 (24%), Positives = 38/85 (44%), Gaps = 6/85 (7%)
Frame = -1
Query: 89 PFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDI------LVFKQED 142
P + E W+ Q +WE K + P T+ R + ND+ +V E+
Sbjct: 262 PINEASRREMWVRKQ---KCVSWE*KPTKDERSEMPETIRRAMPNDVGGIFRRIVGANEE 92
Query: 143 TENLHEALERFKKLLRKCPQHNLTL 167
E++ +E+FK+L R+ + + L
Sbjct: 91 AESM--KIEKFKQLRRRLVKEHALL 23
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,997,692
Number of Sequences: 63676
Number of extensions: 324350
Number of successful extensions: 2047
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 2001
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2040
length of query: 705
length of database: 12,639,632
effective HSP length: 104
effective length of query: 601
effective length of database: 6,017,328
effective search space: 3616414128
effective search space used: 3616414128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0103.9


