

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.7
(411 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
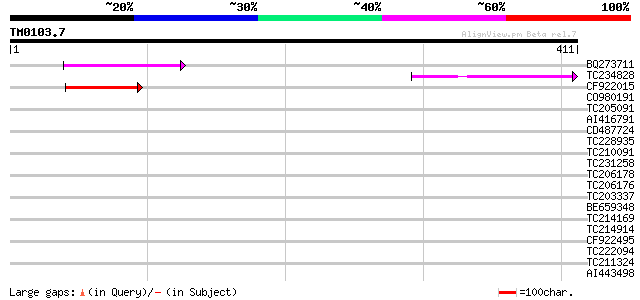
Score E
Sequences producing significant alignments: (bits) Value
BQ273711 89 3e-18
TC234828 80 2e-15
CF922015 72 3e-13
CO980191 39 0.003
TC205091 weakly similar to UP|Q6TXI7 (Q6TXI7) LRRGT00012, partia... 37 0.021
AI416791 33 0.18
CD487724 32 0.40
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 32 0.68
TC210091 similar to UP|Q9LT69 (Q9LT69) Phosphoglycerate dehydrog... 31 0.89
TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kin... 30 1.5
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 30 1.5
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 30 1.5
TC203337 similar to UP|Q9S728 (Q9S728) En/Spm-like transposon pr... 30 1.5
BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor p... 30 1.5
TC214169 similar to UP|Q8LF25 (Q8LF25) DNA-damage inducible prot... 30 2.6
TC214914 homologue to UP|Q8LK71 (Q8LK71) Extensin-like protein, ... 30 2.6
CF922495 30 2.6
TC222094 29 3.4
TC211324 28 5.7
AI443498 28 5.7
>BQ273711
Length = 409
Score = 89.4 bits (220), Expect = 3e-18
Identities = 39/88 (44%), Positives = 51/88 (57%)
Frame = -2
Query: 40 YHGLDKFLKRSPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTW 99
Y GL F K PPKF G Y+P+GA W+ E E+IFE + C E KV +A+++L EA W
Sbjct: 264 YRGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAMGCLEEHKVLYATFMLQGEAENW 85
Query: 100 WTSVRGRITPEEGELIWEIFKNSFLEKY 127
W V+ G + W FK+ FLE Y
Sbjct: 84 WKFVKPSFVAPGGVIPWNAFKDKFLENY 1
>TC234828
Length = 857
Score = 80.1 bits (196), Expect = 2e-15
Identities = 48/120 (40%), Positives = 63/120 (52%)
Frame = +3
Query: 292 VLGGQSGVQTGGSGNPNPTTTVGGGSNRNANVPPRDNRRVGHPANKGKVFAMTMEEASES 351
V G + + N PT + G+ N N+ G P +VFAM+ EA+ S
Sbjct: 9 VPGRPTETANSNTDNRGPTNSTRSGNVSNNNISG------GRPKVPSRVFAMSGSEAAAS 170
Query: 352 PELIKGMCYVNGFPLKVLYDSGATHSFISHERVKQLGFPVTSLA*DLEISTPTKNTSRTS 411
+LI+G C + L VLYDSGATHSFISH V++LG T L D+ +STPT TS
Sbjct: 171 DDLIRGKCLIADKLLDVLYDSGATHSFISHACVERLGLCATELPYDMVVSTPTSEPVTTS 350
>CF922015
Length = 172
Score = 72.4 bits (176), Expect = 3e-13
Identities = 28/56 (50%), Positives = 44/56 (78%)
Frame = -3
Query: 41 HGLDKFLKRSPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEA 96
HGLD+F + +PP F+GGY+P+GA W+RE+E+IF + C + +KV FA+++L+ EA
Sbjct: 170 HGLDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFRVMECQDHQKVLFATHMLADEA 3
>CO980191
Length = 802
Score = 39.3 bits (90), Expect = 0.003
Identities = 20/44 (45%), Positives = 24/44 (54%)
Frame = +1
Query: 117 EIFKNSFLEKYFPADAKCRKEMEFFELKQEAMSVGEYAAKFEEL 160
E+FK FLEK+ D K+ EF ELK M V Y F+EL
Sbjct: 373 EVFKEIFLEKHLHEDI*NHKKTEFLELKHGNMIVANYLVNFDEL 504
>TC205091 weakly similar to UP|Q6TXI7 (Q6TXI7) LRRGT00012, partial (5%)
Length = 1360
Score = 36.6 bits (83), Expect = 0.021
Identities = 18/54 (33%), Positives = 27/54 (49%)
Frame = +1
Query: 50 SPPKFEGGYNPDGAYEWVRELERIFETLVCAEPRKVAFASYLLSSEARTWWTSV 103
S P F G N + +W ++E++F +E RKV A+ A WWTS+
Sbjct: 1135 SLPYFHGKDNVEAYLDWEMKVEQLFACHHISEERKVPLATLSFQGYALYWWTSL 1296
>AI416791
Length = 420
Score = 33.5 bits (75), Expect = 0.18
Identities = 37/127 (29%), Positives = 58/127 (45%), Gaps = 5/127 (3%)
Frame = -3
Query: 110 EEGELIWEIFKNSFLEKY-FPADAKCRKEMEFFELKQEAMSVGEYAAKFEELCQFHPRYS 168
E+ +L WE K + LE+Y D +++ ELKQE SV +Y +FE L PR
Sbjct: 391 EDEDLTWESLKIALLERYGGHGDGDVYEQLT--ELKQEG-SVEDYITEFEYLIAQIPRL- 224
Query: 169 TANDETSKCVKFEYGLRPDIRTVVGHDQIRNFATL--VKKCRIFEENEKT*KEYL--KGL 224
E F +GL+ +IR ++R AT+ + + ++ + KE G
Sbjct: 223 ---PEKQFQGYFLHGLQSEIR-----GRVRTLATMGEMSRTKLLQVTRAVEKEVKGGNGS 68
Query: 225 GVNKAPK 231
G + PK
Sbjct: 67 GFGRGPK 47
>CD487724
Length = 676
Score = 32.3 bits (72), Expect = 0.40
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = +3
Query: 365 PLKVLYDSGATHSFISHERVKQLGFPVTS 393
PL L D G+TH+F+ V QLG P S
Sbjct: 21 PLVYLVDGGSTHNFVQQPLVSQLGLPCRS 107
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 31.6 bits (70), Expect = 0.68
Identities = 13/20 (65%), Positives = 14/20 (70%)
Frame = +1
Query: 276 CGRCRRNGHRAQECIVVLGG 295
C RCRR GHRA+ C VL G
Sbjct: 346 CLRCRRRGHRAKNCPEVLDG 405
>TC210091 similar to UP|Q9LT69 (Q9LT69) Phosphoglycerate dehydrogenase,
partial (36%)
Length = 786
Score = 31.2 bits (69), Expect = 0.89
Identities = 22/56 (39%), Positives = 27/56 (47%)
Frame = -3
Query: 262 GFQGRPGGFNNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTTVGGGS 317
GF GR G ++ P GH +EC VLGG GV GG + +GGGS
Sbjct: 562 GFDGRIG-LSDVP--------GHGCKECDSVLGGGDGV--GGGRIDDQAPVLGGGS 428
>TC231258 similar to UP|Q886T6 (Q886T6) Sensory box histidine kinase/response
regulator, partial (3%)
Length = 1374
Score = 30.4 bits (67), Expect = 1.5
Identities = 16/53 (30%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = +2
Query: 116 WEIFKNSFLEKYFPADAKCRKEMEFFELKQ-EAMSVGEYAAKFEELCQFHPRY 167
W+ K FLEK+FPA + ++Q S+ EY +F++ C P +
Sbjct: 734 WDDLKRVFLEKFFPASRTTTIRKDISGIRQLSRESLYEYWERFKKSCASCPHH 892
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 30.4 bits (67), Expect = 1.5
Identities = 23/83 (27%), Positives = 32/83 (37%), Gaps = 21/83 (25%)
Frame = +2
Query: 272 NTPFCGRCRRNGHRAQEC---------------------IVVLGGQSGVQTGGSGNPNPT 310
N C CR+ GH A++C VLG +SG GG G
Sbjct: 638 NEKACNNCRKTGHLARDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGG----- 802
Query: 311 TTVGGGSNRNANVPPRDNRRVGH 333
GGG +V R+ +++GH
Sbjct: 803 -ARGGGGGGYRDVVCRNCQQLGH 868
Score = 28.5 bits (62), Expect = 5.7
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = +2
Query: 269 GFNNTPFCGRCRRNGHRAQEC 289
GF+ C C+R GH A+EC
Sbjct: 323 GFSRDNLCKNCKRPGHYAREC 385
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 30.4 bits (67), Expect = 1.5
Identities = 21/61 (34%), Positives = 26/61 (42%), Gaps = 3/61 (4%)
Frame = +3
Query: 259 GGFGFQGRPGG---FNNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTTVGG 315
GG+G G GG C C +GH A++C GG G + GG G GG
Sbjct: 375 GGYGGGGGYGGGGRGGGGGACYNCGESGHLARDC---SGGGGGDRYGGGGGGGRYGGGGG 545
Query: 316 G 316
G
Sbjct: 546 G 548
>TC203337 similar to UP|Q9S728 (Q9S728) En/Spm-like transposon protein
(Protodermal factor 1), partial (46%)
Length = 1410
Score = 30.4 bits (67), Expect = 1.5
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = +2
Query: 302 GGSGNPNPTTTVGGGSNRNANVPPRD 327
GGS NP P+T GGGS PP D
Sbjct: 428 GGSYNPTPSTPPGGGSYDPTPSPPTD 505
>BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor protein
DSK-R1 - fruit fly (Drosophila melanogaster), partial
(6%)
Length = 770
Score = 30.4 bits (67), Expect = 1.5
Identities = 14/36 (38%), Positives = 16/36 (43%)
Frame = -1
Query: 259 GGFGFQGRPGGFNNTPFCGRCRRNGHRAQECIVVLG 294
GG G QG G P C C+R GH C + G
Sbjct: 434 GGRGNQGGRAGRGGRP*CSYCKRVGHTQDTCYSIHG 327
>TC214169 similar to UP|Q8LF25 (Q8LF25) DNA-damage inducible protein
DDI1-like, partial (90%)
Length = 1604
Score = 29.6 bits (65), Expect = 2.6
Identities = 14/28 (50%), Positives = 17/28 (60%)
Frame = +2
Query: 361 VNGFPLKVLYDSGATHSFISHERVKQLG 388
VNG PLK DSGA + IS ++LG
Sbjct: 764 VNGVPLKAFVDSGAQSTIISKSCAERLG 847
>TC214914 homologue to UP|Q8LK71 (Q8LK71) Extensin-like protein, complete
Length = 852
Score = 29.6 bits (65), Expect = 2.6
Identities = 31/112 (27%), Positives = 42/112 (36%)
Frame = +3
Query: 225 GVNKAPKKKEEKRKPYF*PQDQGRNQFGRTWNPTGGFGFQGRPGGFNNTPFCGRCRRNGH 284
G +PK + K KP +P+GG G G GG + G +G
Sbjct: 192 GTCPSPKPRHNKHKPV-------------KPSPSGGSGGSGGSGGSGGSGGSGGSGGSGG 332
Query: 285 RAQECIVVLGGQSGVQTGGSGNPNPTTTVGGGSNRNANVPPRDNRRVGHPAN 336
GG G +GGSG GGS ++ PRD ++G AN
Sbjct: 333 S--------GGSGG--SGGSGG-------SGGSGGSSTSCPRDALKLGVCAN 437
>CF922495
Length = 427
Score = 29.6 bits (65), Expect = 2.6
Identities = 30/115 (26%), Positives = 48/115 (41%), Gaps = 1/115 (0%)
Frame = +3
Query: 228 KAPKKKEEKRKPYF*PQDQGRNQFGRTWNPTGGFGFQGRP-GGFNNTPFCGRCRRNGHRA 286
K PKKK +K P +G + +P GG + P GG G ++ G
Sbjct: 45 KTPKKKISPKKKRGPPPPKGFPGKPKGGHPGGGPPQKSPPWGGPKREKPGGNFKKGGAGG 224
Query: 287 QECIVVLGGQSGVQTGGSGNPNPTTTVGGGSNRNANVPPRDNRRVGHPANKGKVF 341
++ GG++ + GG +GGG +N PR +R G + +G+ F
Sbjct: 225 KKP----GGENLKKGGGGKTKILAGPLGGGGKKNGAKKPRGEKR-GPNSPRGQFF 374
>TC222094
Length = 984
Score = 29.3 bits (64), Expect = 3.4
Identities = 15/40 (37%), Positives = 19/40 (47%)
Frame = -2
Query: 90 YLLSSEARTWWTSVRGRITPEEGELIWEIFKNSFLEKYFP 129
+ L+ EARTW S +G W FL+KYFP
Sbjct: 605 FSLAGEARTWLRSFKG-----NNLRTWNEXXEKFLKKYFP 501
>TC211324
Length = 845
Score = 28.5 bits (62), Expect = 5.7
Identities = 12/36 (33%), Positives = 18/36 (49%)
Frame = +1
Query: 26 QGAGQGAANNQEEIYHGLDKFLKRSPPKFEGGYNPD 61
+G GA +E +H DK+L S P ++PD
Sbjct: 382 KGMVNGAVETCKESFHRFDKYLNFSNPLISNNFSPD 489
>AI443498
Length = 377
Score = 28.5 bits (62), Expect = 5.7
Identities = 25/76 (32%), Positives = 30/76 (38%), Gaps = 5/76 (6%)
Frame = -3
Query: 255 WNPTGGFGFQGRPGGF--NNTPFCGRCRRNGHRAQECIVVLGGQSGVQTGGSGNPNPTTT 312
W P GGF + + GGF P G R G VVLGG+ + GG P
Sbjct: 237 WGPGGGFFLKKKGGGFTKGGKPLPGTPPRGG-------VVLGGKK--KRGGVWVPKGFFF 85
Query: 313 VGG---GSNRNANVPP 325
+GG G PP
Sbjct: 84 LGGKGIGGGGGKEFPP 37
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,711,434
Number of Sequences: 63676
Number of extensions: 260970
Number of successful extensions: 1374
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1338
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1372
length of query: 411
length of database: 12,639,632
effective HSP length: 100
effective length of query: 311
effective length of database: 6,272,032
effective search space: 1950601952
effective search space used: 1950601952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0103.7


