

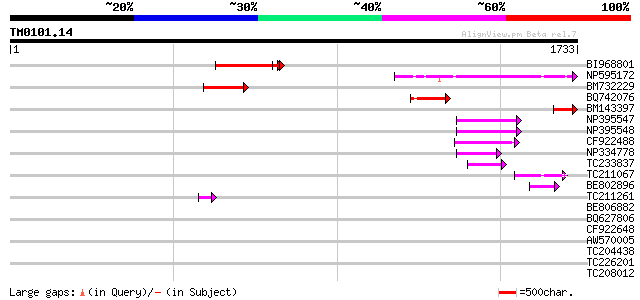
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0101.14
(1733 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI968801 similar to GP|24461860|gb CTV.20 {Poncirus trifoliata},... 259 9e-69
NP595172 polyprotein [Glycine max] 230 4e-60
BM732229 190 5e-48
BQ742076 134 5e-31
BM143397 119 1e-26
NP395547 reverse transcriptase [Glycine max] 92 2e-18
NP395548 reverse transcriptase [Glycine max] 82 3e-15
CF922488 75 3e-13
NP334778 reverse transcriptase [Glycine max] 70 8e-12
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 60 6e-09
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 57 9e-08
BE802896 53 1e-06
TC211261 47 6e-05
BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, pa... 42 0.002
BQ627806 42 0.002
CF922648 40 0.009
AW570005 39 0.020
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 38 0.044
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 37 0.076
TC208012 similar to UP|Q761Z7 (Q761Z7) BRI1-KD interacting prote... 37 0.076
>BI968801 similar to GP|24461860|gb CTV.20 {Poncirus trifoliata}, partial
(0%)
Length = 660
Score = 259 bits (661), Expect = 9e-69
Identities = 129/206 (62%), Positives = 160/206 (77%), Gaps = 1/206 (0%)
Frame = -1
Query: 629 NMTMVATAYETSNNCPEYLIVEILVADFCGQLKGWWDNYLTEDEKNQILTAVKTDEEGNP 688
+MTMVATAY+TS+ C I++ILVA F GQLK WWDNYLT +EK +I +AVKTD G
Sbjct: 657 HMTMVATAYQTSHECSXXXIIDILVAGFSGQLKRWWDNYLTNEEK*KIYSAVKTDLNGKV 478
Query: 689 II-EDGKLISDAVNSLIFTIAQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTR 747
I +D K I DAVN+LIFTIAQHF+GDPSL KDRS +LLSNLKC++L DFRWY+DTFLTR
Sbjct: 477 ITNDDDKEILDAVNTLIFTIAQHFIGDPSLWKDRSAELLSNLKCRTLADFRWYRDTFLTR 298
Query: 748 VYTREDSQQAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQLIAIIQRVAL 807
VYTREDSQQ F KEKFLAGLP+S GDKVR+K+RSQ+ G+IPY++LSYGQLI+ +Q+VA
Sbjct: 297 VYTREDSQQPF*KEKFLAGLPRSLGDKVRDKIRSQSANGDIPYESLSYGQLISYVQKVAX 118
Query: 808 KICQDDKIQQQLTKEKSQNRRDLGTF 833
K + + +++ K N+ F
Sbjct: 117 KNLSG*QNSKAISQRKGSNKEGFRFF 40
Score = 58.2 bits (139), Expect = 3e-08
Identities = 25/36 (69%), Positives = 31/36 (85%)
Frame = -2
Query: 803 QRVALKICQDDKIQQQLTKEKSQNRRDLGTFCEQFG 838
+R KICQDDKIQ+QL KEK+Q ++DLG+FCEQFG
Sbjct: 131 KR*XXKICQDDKIQRQLAKEKAQTKKDLGSFCEQFG 24
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 230 bits (587), Expect = 4e-60
Identities = 172/572 (30%), Positives = 279/572 (48%), Gaps = 15/572 (2%)
Frame = +1
Query: 1177 EIETPFLLVRNLTHKVIIGTPFIKKLFPYNTDEKGITVQHLGKPILFKFSKPPFSKTLNI 1236
E++ P L++ VI+G+ ++ L P+ D +T++ S+
Sbjct: 1378 EVKVPVYLLQISGADVILGSTWLATLGPHVADYAALTLKFFQNDKFITLQGEGNSEAT-- 1551
Query: 1237 ISYKEKQINFLKEEISHKSIE----VQLQQPSVKTRIGNILENIQSSICSDLP---NAFW 1289
+ Q++ + + KSIE +QL Q V + L+++ ++I +L + +
Sbjct: 1552 ----QAQLHHFRRLQNTKSIEECFAIQLIQKEVPE---DTLKDLPTNIDPELAILLHTYA 1710
Query: 1290 ERKSHMVELPYEKDFSDKQIP-------TKARPIQMNEELLQFCQKEINDLLQKKLIRRS 1342
+ + LP +++ D IP K RP + +K I ++L + +I+ S
Sbjct: 1711 QVFAVPASLPPQRE-QDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPS 1887
Query: 1343 KSPWSCATFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSK 1402
SP+S V K+ G+ R +Y+ LN +P+P +LL LH A+ FSK
Sbjct: 1888 NSPFSLPILLVKKKD----GSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSK 2055
Query: 1403 FDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFN-PYSNF 1461
D++SG+ QI +Q +DR KTAF G YEW VMPFGL NAP+ FQ +MN+IF F
Sbjct: 2056 LDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKF 2235
Query: 1462 TIVYIDDVLIFSQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTI 1521
+V+ DD+LI+S S H KHL + + +K++ L +K S T++ +LGH + +
Sbjct: 2236 VLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGV 2415
Query: 1522 IPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPPPWSD 1581
N ++ +P K QL+ FLG Y F + I L D L+KD W++
Sbjct: 2416 SMENTKVQAVLDWPTPNNVK-QLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKDSFLWNN 2592
Query: 1582 VHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKH 1641
++K + P L LP+ I+ETDAS IG G +L Q N IA+ SK
Sbjct: 2593 EAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQ----NGHPIAYFSKK 2760
Query: 1642 WNPAQQNYSTVKKEVLAIVLSISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIF 1701
P Q S +E+LAI ++SKF++ L+ F++R D +S K ++ + ++ +
Sbjct: 2761 LAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQ--- 2931
Query: 1702 ARWQAILSVFDFEIEYIKGSTNSLPDFLTREY 1733
W +DF+IEY G N D L+R +
Sbjct: 2932 -AWLHKFLGYDFKIEYKPGKDNQAADALSRMF 3024
>BM732229
Length = 421
Score = 190 bits (482), Expect = 5e-48
Identities = 94/139 (67%), Positives = 111/139 (79%), Gaps = 1/139 (0%)
Frame = -3
Query: 591 DLLLEEKDSSNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATAYETSNNCPEYLIVE 650
DLLLEE+ +NFKSFSANN+YEWNID + EY I LQ+MTMVATAY+TS+ C E I++
Sbjct: 419 DLLLEERGENNFKSFSANNIYEWNIDAQTEYNIMNTLQHMTMVATAYQTSHECSEETIID 240
Query: 651 ILVADFCGQLKGWWDNYLTEDEKNQILTAVKTDEEGNPII-EDGKLISDAVNSLIFTIAQ 709
ILVA F GQLKGWWDNYLT +EK++I +AVKTD G I +D K I DAVN+LIFTIAQ
Sbjct: 239 ILVAGFSGQLKGWWDNYLTNEEKSKIYSAVKTDLNGKVITNDDDKEIPDAVNTLIFTIAQ 60
Query: 710 HFVGDPSLIKDRSGDLLSN 728
HF+GDPSL DRS +LLSN
Sbjct: 59 HFIGDPSLW*DRSAELLSN 3
>BQ742076
Length = 429
Score = 134 bits (336), Expect = 5e-31
Identities = 66/120 (55%), Positives = 89/120 (74%)
Frame = -2
Query: 1226 SKPPFSKTLNIISYKEKQINFLKEEISHKSIEVQLQQPSVKTRIGNILENIQSSICSDLP 1285
SK P K +N QINFLK+E+S +I++QL++P +K +I ++L++IQS+IC DL
Sbjct: 344 SKLPSKKKIN-------QINFLKDEVSFSNIQIQLEKPQMKEKISSLLKHIQSTICFDLA 186
Query: 1286 NAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFCQKEINDLLQKKLIRRSKSP 1345
AF RK H++ LP +KDF +K IPTK+RPI MNEELLQ+CQKEI DL K LIR+SK+P
Sbjct: 185 RAF*NRKKHILNLPCDKDFKEKHIPTKSRPI*MNEELLQYCQKEIKDLFNKGLIRKSKNP 6
>BM143397
Length = 408
Score = 119 bits (298), Expect = 1e-26
Identities = 57/71 (80%), Positives = 64/71 (89%)
Frame = -3
Query: 1663 ISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGST 1722
ISKFQ DL+NQ L+RVDCK AKDILQKDVKNLASK IFARWQAILSVFDF+IEY K ++
Sbjct: 406 ISKFQSDLLNQKILIRVDCKLAKDILQKDVKNLASKQIFARWQAILSVFDFDIEYFKDTS 227
Query: 1723 NSLPDFLTREY 1733
NSLPD+LTRE+
Sbjct: 226 NSLPDYLTREF 194
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 92.0 bits (227), Expect = 2e-18
Identities = 61/201 (30%), Positives = 95/201 (46%), Gaps = 1/201 (0%)
Frame = +1
Query: 1365 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1424
R+ I+Y+ LN+A YP+P +L RL + D SG+ QI + +D+ KTAF
Sbjct: 163 RMCIDYRKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAF 342
Query: 1425 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQHFKHL 1483
T PF + + MPFGL NA + FQR M IF+ I V++DD F S +L
Sbjct: 343 TCPFSVFAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANL 522
Query: 1484 NTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQ 1543
+ +K+ L ++ K + LGH I + I + ++ DK P + K
Sbjct: 523 EKVLQRCEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVK-G 699
Query: 1544 LQRFLGCLNYVADFCPQLSTI 1564
+ FLG + + F + +
Sbjct: 700 IHSFLGHVGFYRRFIKDFTKV 762
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 81.6 bits (200), Expect = 3e-15
Identities = 59/201 (29%), Positives = 92/201 (45%), Gaps = 1/201 (0%)
Frame = +1
Query: 1365 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1424
+L I+Y+ LN+A +P+P +L RL + D G+ QI + KD+ K AF
Sbjct: 163 KLCIDYRKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAF 342
Query: 1425 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQHFKHL 1483
T PFG + + +PFGL NAP+ FQ M IF +I V++DD +F S++ K L
Sbjct: 343 TCPFGVFAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKL 522
Query: 1484 NTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQ 1543
+ + L ++ K + LGH I I I+ +K P K
Sbjct: 523 EMVLQRCVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVK-G 699
Query: 1544 LQRFLGCLNYVADFCPQLSTI 1564
++ FLG + F + +
Sbjct: 700 IRSFLGQARFYRRFIKDFTKV 762
>CF922488
Length = 741
Score = 74.7 bits (182), Expect = 3e-13
Identities = 58/201 (28%), Positives = 100/201 (48%), Gaps = 3/201 (1%)
Frame = +3
Query: 1360 ERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDR 1419
E G + ++Y+ LN A ++P+P+ L+ FS D SG+ QI++ +D
Sbjct: 12 EDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAPEDM 191
Query: 1420 YKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQ 1478
KT F +G + + M FGLKN + +QR M +F + I VY+DD+++ S++ ++
Sbjct: 192 EKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRTEEE 371
Query: 1479 HFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIH--QGTIIPINRAIEFTDKFPD 1536
H +L ++K L ++ K +F+ K R L I +G + N+ +
Sbjct: 372 HLVNLRKLFRRLRKYRLRLNPAK-CMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMAKP 548
Query: 1537 QIIDKTQLQRFLGCLNYVADF 1557
+ Q+Q FLG LNY+ F
Sbjct: 549 H--TEKQVQGFLGRLNYIVRF 605
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 70.1 bits (170), Expect = 8e-12
Identities = 40/138 (28%), Positives = 74/138 (52%), Gaps = 1/138 (0%)
Frame = +3
Query: 1365 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1424
R+ ++Y+ LN+A +P+P+ L+A + +FS D SG+ QI++ +D KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 1425 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPYSNFTI-VYIDDVLIFSQSIDQHFKHL 1483
+G + + VM FGLKN + + R M +F + I Y+D+++ S+ ++H +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 1484 NTFISVIKKNGLAVSKTK 1501
++K L ++ K
Sbjct: 363 QNLFGQLRKYRLRLNPRK 416
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 60.5 bits (145), Expect = 6e-09
Identities = 38/120 (31%), Positives = 63/120 (51%), Gaps = 1/120 (0%)
Frame = +2
Query: 1400 FSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNPYS 1459
FS D SG+ QI + +D KT F +G + + VM FGLKN + +QR M +F+
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 1460 NFTI-VYIDDVLIFSQSIDQHFKHLNTFISVIKKNGLAVSKTKVSLFQTKIRFLGHNIHQ 1518
+ I VY+DD++ S++ +H +L ++K L ++ TK + + LG + Q
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 56.6 bits (135), Expect = 9e-08
Identities = 43/162 (26%), Positives = 77/162 (46%), Gaps = 1/162 (0%)
Frame = +1
Query: 1544 LQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPP-PWSDVHTNVVKQIKLRIKNLPCLYL 1602
++ F G ++ F P ST+ L++ +KK+ W + +K ++ P L L
Sbjct: 121 IRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKAPVLAL 300
Query: 1603 PNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLS 1662
P+ +E DAS +G +L Q IA+ S+ + A NY T KE+ A++ +
Sbjct: 301 PDFSKTFELECDASGVGVRAVLLQ----GGHPIAYFSEKLHSATLNYPTYDKELYALIRA 468
Query: 1663 ISKFQYDLINQTFLVRVDCKSAKDILQKDVKNLASKHIFARW 1704
+++ L+ + F++ D +S K I K L +H A+W
Sbjct: 469 PQTWEHFLVCKEFVIHSDHQSLKYIRGK--SKLNKRH--AKW 582
>BE802896
Length = 416
Score = 52.8 bits (125), Expect = 1e-06
Identities = 30/91 (32%), Positives = 51/91 (55%)
Frame = -2
Query: 1590 IKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNY 1649
+K + P + P+ A + DAS+ G +L QKI ++I ++S+ + AQ NY
Sbjct: 280 LKRALITTPIIQAPDWTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANY 101
Query: 1650 STVKKEVLAIVLSISKFQYDLINQTFLVRVD 1680
+T +KE+LAIV ++ KF L+ +V +D
Sbjct: 100 TTTEKELLAIVFALEKFHSYLLGTRIIVYID 8
>TC211261
Length = 520
Score = 47.4 bits (111), Expect = 6e-05
Identities = 25/55 (45%), Positives = 33/55 (59%), Gaps = 1/55 (1%)
Frame = -2
Query: 577 RKETKLYYPRATAPDLLLEEKD-SSNFKSFSANNVYEWNIDGENEYGITKILQNM 630
R TKLYY + T PDL L+E+D +S + F + NIDG+ EY T LQ+M
Sbjct: 504 RNSTKLYYQKVTIPDLWLQERDLNSKLQRFRTMGIC**NIDGQIEYNTTHTLQHM 340
>BE806882 similar to GP|27764548|gb polyprotein {Glycine max}, partial (3%)
Length = 153
Score = 42.4 bits (98), Expect = 0.002
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Frame = -1
Query: 1440 LKNAPSEFQRIMNEIF-NPYSNFTIVYIDDVLIFSQSIDQHFKHLNTFISV 1489
L NAP+ FQ +MN IF + + +V+ DD+L++S + +H HL V
Sbjct: 153 LTNAPTSFQCLMNHIFQHALRKYVLVFFDDILVYSSTWHEHLCHLEVVFKV 1
>BQ627806
Length = 435
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/55 (41%), Positives = 30/55 (53%)
Frame = +3
Query: 1572 LKKDPPPWSDVHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQ 1626
L KD W++ Q+KL + P L LP+ + +VETDAS IG G IL Q
Sbjct: 6 LVKDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQ 170
>CF922648
Length = 570
Score = 40.0 bits (92), Expect = 0.009
Identities = 29/94 (30%), Positives = 44/94 (45%), Gaps = 7/94 (7%)
Frame = +3
Query: 821 KEKSQNRRDLGTFCEQFGIQGCPKK----PKPRKHDPPPKQQWRRNSSRNHDHRKPKPRS 876
+E +++LG + G+ G PK P PR H PPK+ + +N R PK S
Sbjct: 237 RESKTPQKNLGNLPK--GLWGPPKTGGGTPPPRGHQIPPKKPGVKGG*KN---RGPKKNS 401
Query: 877 KPHSTQAAKNPPENRPSQ---GKNVTCYNCGKPG 907
+ + PP+N P + GKN ++ G PG
Sbjct: 402 QKPPQRGQPGPPKNPPPEKIRGKNPPKFSPGNPG 503
>AW570005
Length = 413
Score = 38.9 bits (89), Expect = 0.020
Identities = 37/128 (28%), Positives = 53/128 (40%)
Frame = -2
Query: 1544 LQRFLGCLNYVADFCPQLSTIIKLLHDRLKKDPPPWSDVHTNVVKQIKLRIKNLPCLYLP 1603
L+ FL + F + + L L KD WS + +K + N L LP
Sbjct: 388 LRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDSFVWSPEADVAFQALKNVVTNTLVLALP 209
Query: 1604 NPQAFKIVETDASDIGFGGILKQKIFDNEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSI 1663
+ VETDAS G +L Q+ IAF SK + P ST E+ AI +
Sbjct: 208 DFTKPFTVETDASGSDMGAVLSQE----GHPIAFFSKEFCPKLVRSSTYVHELAAITNVV 41
Query: 1664 SKFQYDLI 1671
K++ L+
Sbjct: 40 KKWRQYLL 17
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 37.7 bits (86), Expect = 0.044
Identities = 27/114 (23%), Positives = 51/114 (44%), Gaps = 15/114 (13%)
Frame = +1
Query: 857 QQWRRNSSRNHDHRKPKPRSKPHSTQAA---KNPPENRPSQGKNVTCYNCGKPGHISRYC 913
+Q+ + +R +KP R+ P + + + +PS K + C+ C GHI C
Sbjct: 766 KQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKKSDEKPSHSKGIQCHGCEGYGHIKAEC 945
Query: 914 --RLKRR----------ISELHLEPEIEDKINNLLIQTSDEEESASSDSEVSED 955
LK++ +E E + + +N L + E+S+ +DSE++ D
Sbjct: 946 PTHLKKQRKGLSVCRSDDTESEQESDSDRDVNALTGRFESAEDSSDTDSEITFD 1107
Score = 31.6 bits (70), Expect = 3.2
Identities = 16/39 (41%), Positives = 20/39 (51%)
Frame = +1
Query: 875 RSKPHSTQAAKNPPENRPSQGKNVTCYNCGKPGHISRYC 913
RS+ H TQ K S+ K C+ CGK GHI +C
Sbjct: 1459 RSRHHGTQQKK-------SKRKKWRCHYCGKYGHIKPFC 1554
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 37.0 bits (84), Expect = 0.076
Identities = 12/24 (50%), Positives = 18/24 (75%)
Frame = +1
Query: 893 SQGKNVTCYNCGKPGHISRYCRLK 916
S G ++ CY CG+PGH +R CR++
Sbjct: 226 SGGSDLKCYECGEPGHFARECRMR 297
>TC208012 similar to UP|Q761Z7 (Q761Z7) BRI1-KD interacting protein 117
(Fragment), partial (38%)
Length = 993
Score = 37.0 bits (84), Expect = 0.076
Identities = 49/229 (21%), Positives = 90/229 (38%), Gaps = 11/229 (4%)
Frame = +1
Query: 871 KPKPRSKPHSTQAAKNPPENRPSQGKNVT------CYNCGKPGHISRYCRLKRRISELHL 924
K + S + T P + PS V CY C K GH+SR C+ ++ LH
Sbjct: 31 KKEETSSENDTLDQGKKPGSGPSDAPKVPSDAPKICYKCKKAGHLSRDCK-EQPDGLLHR 207
Query: 925 EP--EIEDKINNLLIQTSDEEESASSDSEVSEDLNQIQNDDDPQSSSSINVLTNE---QD 979
E E+ + I TS + A + +D+N+I +++ + + ++ LT D
Sbjct: 208 NAIGEAEENPKSTAIDTSQADRVAMEE----DDINEI-GEEEKEKLNDVDYLTGNPLPND 372
Query: 980 LLFRAINSIPDPDEKKIYLERLKFTLEDKPPKNPITTNKFNLRDTFRRLEKSTIKPVTIQ 1039
+L A+ + Y R+K + K NL F + ++T + +
Sbjct: 373 ILLYAVPVCGPYSAVQSYKYRVKI-IPGPAKKGKAAKTAMNL---FSHMSEATTREKELM 540
Query: 1040 DLQSEVHTLQAEVKSLKQIQISQQLILDKLTEENSEESSSSSSTPNSAS 1088
++ + A V ++K + K ++ ++SS S P S
Sbjct: 541 KACTDPELVAAIVGNVKISAAGLTQLKQK--QKKGKKSSKQES*PTHGS 681
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.339 0.149 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 81,940,250
Number of Sequences: 63676
Number of extensions: 1286560
Number of successful extensions: 13167
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 11446
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12833
length of query: 1733
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1623
effective length of database: 5,635,272
effective search space: 9146046456
effective search space used: 9146046456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0101.14


