

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100c.1
(773 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
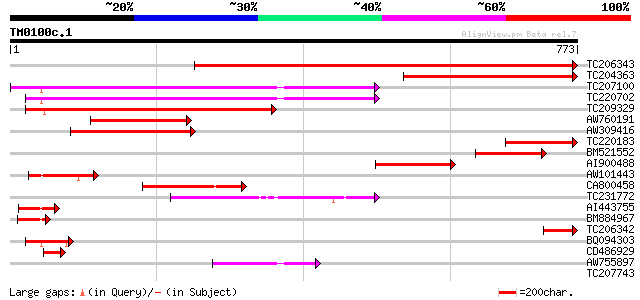
Score E
Sequences producing significant alignments: (bits) Value
TC206343 similar to UP|O22403 (O22403) ADL2 protein, partial (53%) 880 0.0
TC204363 similar to UP|O22403 (O22403) ADL2 protein, partial (15%) 378 e-105
TC207100 UP|Q39828 (Q39828) SDL5A, complete 329 2e-90
TC220702 UP|Q39821 (Q39821) SDL12A, complete 328 7e-90
TC209329 similar to UP|Q9FNX5 (Q9FNX5) Dynamin-like protein DLP2... 280 2e-75
AW760191 255 5e-68
AW309416 251 9e-67
TC220183 weakly similar to UP|Q8LPH8 (Q8LPH8) Dynamin-like prote... 155 5e-38
BM521552 similar to PIR|T04982|T049 dynamin-like protein ADL2 - ... 154 1e-37
AI900488 144 1e-34
AW101443 128 1e-29
CA800458 116 4e-26
TC231772 similar to UP|Q9FNX5 (Q9FNX5) Dynamin-like protein DLP2... 104 2e-22
AI443755 similar to GP|19032339|dbj dynamin like protein 2b {Ara... 86 8e-17
BM884967 71 2e-12
TC206342 67 4e-11
BQ094303 PIR|S63667|S63 phragmoplastin 12 - soybean, partial (13%) 62 1e-09
CD486929 54 3e-07
AW755897 homologue to PIR|S63667|S63 phragmoplastin 12 - soybean... 53 6e-07
TC207743 similar to UP|Q8S3C8 (Q8S3C8) Dynamin-like protein C, p... 41 0.002
>TC206343 similar to UP|O22403 (O22403) ADL2 protein, partial (53%)
Length = 1749
Score = 880 bits (2273), Expect = 0.0
Identities = 451/526 (85%), Positives = 482/526 (90%), Gaps = 5/526 (0%)
Frame = +1
Query: 253 VNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLP 312
VNRSQEDI MNRSIKDALVAEE+FFR+ P+YSGLADSCGVPQLAKKLNKILAQHIK+VLP
Sbjct: 1 VNRSQEDILMNRSIKDALVAEEKFFRTHPIYSGLADSCGVPQLAKKLNKILAQHIKSVLP 180
Query: 313 GLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCE 372
GL+ARIS SLV +AKEHASYGEITESKAGQGAL+LNILSKYCDAFSSM+EGKN+EMST E
Sbjct: 181 GLRARISASLVTIAKEHASYGEITESKAGQGALLLNILSKYCDAFSSMVEGKNEEMSTSE 360
Query: 373 LSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRR 432
LSGGARIHYIFQSI+V+SLE VDPCEDLTDDDIRTA+QNATGPKSALF+PEVPFE+ VRR
Sbjct: 361 LSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPKSALFVPEVPFEVLVRR 540
Query: 433 QISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETM 492
QISRLLDPSLQCARFIYDELMKISH+CM TELQRFPFLRK MDEVIGNFLR+GLEPSETM
Sbjct: 541 QISRLLDPSLQCARFIYDELMKISHQCMVTELQRFPFLRKRMDEVIGNFLREGLEPSETM 720
Query: 493 ITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LDALESDKGLASERN 550
ITH+IEMEMDYINTSHPNFIGGSKA+EAAVQQT+ SRV PVS DALESDKG A+ER+
Sbjct: 721 ITHVIEMEMDYINTSHPNFIGGSKAIEAAVQQTRSSRVGLPVSRVKDALESDKGPATERS 900
Query: 551 GKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKENTS 610
GKSR I+AR ANGVVAD GVR SDVEKV SGN GGSSW ISSIFGG DNRV VKE T+
Sbjct: 901 GKSRSILARHANGVVADQGVRATSDVEKVVTSGNTGGSSWGISSIFGGGDNRVSVKEMTA 1080
Query: 611 TKPH-AEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYYDIARKNIE 669
+KPH EP+HNVQS STIHLREPP VLRP ESNSE E VEITVTKLLL+SYYDI RKN+E
Sbjct: 1081SKPHTTEPMHNVQSFSTIHLREPPSVLRPLESNSETEAVEITVTKLLLKSYYDIVRKNVE 1260
Query: 670 DLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRELLRAYQQA 729
DLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEP EVA KRK CRELLRAYQQA
Sbjct: 1261DLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPEEVAKKRKNCRELLRAYQQA 1440
Query: 730 FRDLDELPLEAETVERGYGSPE-ATGLPRIRGLPSSSMYS-SSLGD 773
F+DL+ELPLEAETVERGY PE TGLP+IRGLP+SSMYS SSLGD
Sbjct: 1441FKDLNELPLEAETVERGYSLPETTTGLPKIRGLPTSSMYSTSSLGD 1578
>TC204363 similar to UP|O22403 (O22403) ADL2 protein, partial (15%)
Length = 1368
Score = 378 bits (971), Expect = e-105
Identities = 191/239 (79%), Positives = 212/239 (87%), Gaps = 2/239 (0%)
Frame = +3
Query: 537 DALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIF 596
DALESDKG ASER+ KSR I+ARQANGVV D GVR ASDVEK+ PSGN GGSSW ISSIF
Sbjct: 18 DALESDKGSASERSVKSRAILARQANGVVPDPGVRAASDVEKIVPSGNTGGSSWGISSIF 197
Query: 597 GGRDNRVYVKENTSTKPHAEPVHNV-QSSSTIHLREPPPVLRPSESNSEMEGVEITVTKL 655
GG D+R+ VKEN S+KPHAEPVH++ QS S IHLREPPP+LRPSESNSE E +EITVTKL
Sbjct: 198 GGGDSRMSVKENISSKPHAEPVHSIEQSVSMIHLREPPPILRPSESNSETEAIEITVTKL 377
Query: 656 LLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAK 715
LLRSYY I RKN+EDL+PKAIMHFLVNNTKRELHNVFI+KLYR++LFEEMLQEP E+A K
Sbjct: 378 LLRSYYGIVRKNVEDLIPKAIMHFLVNNTKRELHNVFIKKLYRDNLFEEMLQEPDEIAVK 557
Query: 716 RKRCRELLRAYQQAFRDLDELPLEAETVERGYGSPEATGLPRIRGLPSSSMYS-SSLGD 773
RKRCRELLRAYQQAF+DL+ELPLEAETVERGY PE +GLP+I GLP+SSMYS SS GD
Sbjct: 558 RKRCRELLRAYQQAFKDLEELPLEAETVERGYSLPETSGLPKIHGLPTSSMYSTSSSGD 734
>TC207100 UP|Q39828 (Q39828) SDL5A, complete
Length = 2284
Score = 329 bits (844), Expect = 2e-90
Identities = 199/518 (38%), Positives = 303/518 (58%), Gaps = 14/518 (2%)
Frame = +2
Query: 1 MAVTDDEAV--TAASTAATPLG-SSVISLVNRLQDIFSRVGNQS------TIID-LPQVA 50
+A+TD T +S +P+ ++ISLVN++Q + +G+ T+ D LP +A
Sbjct: 86 LALTDRRCYI*TRSSQQISPIQMENLISLVNKIQRACTALGDHGENSALPTLWDSLPAIA 265
Query: 51 VVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFH 110
VVG QSSGKSSVLES+VG+DFLPRG+ I TRRPLVLQL + E+ EFLHLP ++F
Sbjct: 266 VVGGQSSGKSSVLESVVGKDFLPRGSGIVTRRPLVLQLHKIEEGSREYAEFLHLPRKRFT 445
Query: 111 DFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDI 170
DF +R+EIQ ETDRE G K +S I L I+SPNV+++TLVDLPG+TKV V QP I
Sbjct: 446 DFVAVRNEIQDETDRETGRTKQISTVPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPDSI 625
Query: 171 EARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDR 230
I M+ SYI+ P C+ILA++PAN DLA SDA+++++ DP G+RTIGV+TK+D+MD+
Sbjct: 626 VKDIEDMVRSYIEKPNCIILAISPANQDLATSDAIKISREVDPTGDRTIGVLTKIDLMDK 805
Query: 231 GTDARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSC 290
GTDA ++L G+ L+ ++GVVNRSQ+DI N + A E E+F S P Y LA+
Sbjct: 806 GTDAVDILEGRAYRLKFPWIGVVNRSQQDINKNVDMIAARRREREYFNSTPEYKHLANRM 985
Query: 291 GVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGAL--VLN 348
G LAK L+K L IK+ +PG+++ I+ ++ L E G+ + AG G L ++
Sbjct: 986 GSEHLAKMLSKHLETVIKSKIPGIQSLINKTIAELEAELTRLGKPVAADAG-GKLYAIME 1162
Query: 349 ILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTA 408
I + F L+G GG +I+ +F + +L+ + + L+ ++IR
Sbjct: 1163ICRSFDQIFKDHLDGVR--------PGGDKIYNVFDNQLPAALKRLQFDKQLSMENIRKL 1318
Query: 409 LQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCM--FTELQR 466
+ A G + L PE + + + + P+ ++ L + H+ + +L++
Sbjct: 1319ITEADGYQPHLIAPEQGYRRLIESSLITIRGPAEAAVDAVHSLLKDLVHKAISETLDLKQ 1498
Query: 467 FPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYI 504
+P LR + + L + S+ +++ME Y+
Sbjct: 1499YPGLRVEVGAAAVDSLERMRDESKRATLQLVDMECGYL 1612
>TC220702 UP|Q39821 (Q39821) SDL12A, complete
Length = 2213
Score = 328 bits (840), Expect = 7e-90
Identities = 193/494 (39%), Positives = 290/494 (58%), Gaps = 11/494 (2%)
Frame = +1
Query: 22 SVISLVNRLQDIFSRVGNQS------TIID-LPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
++ISLVN++Q + +G+ T+ D LP +AVVG QSSGKSSVLES+VG+DFLPR
Sbjct: 181 NLISLVNKIQRACTALGDHGENSALPTLWDSLPAIAVVGGQSSGKSSVLESVVGKDFLPR 360
Query: 75 GTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVS 134
G+ I TRRPLVLQL E+ EFLHLP ++F DF +R EIQ ETDRE G K +S
Sbjct: 361 GSGIVTRRPLVLQLHKIDEGSREYAEFLHLPRKRFTDFVAVRKEIQDETDRETGRTKQIS 540
Query: 135 EKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTP 194
I L I+SPNV+++TL+DLPG+TKV V QP I I M+ SYI+ P C+ILA++P
Sbjct: 541 SVPIHLSIYSPNVVNLTLIDLPGLTKVAVEGQPDSIVKDIEDMVRSYIEKPNCIILAISP 720
Query: 195 ANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVN 254
AN DLA SDA+++++ DP G+RTIGV+TK+D+MD+GTDA ++L G+ L+ ++GVVN
Sbjct: 721 ANQDLATSDAIKISREVDPTGDRTIGVLTKIDLMDKGTDAVDILEGRAYRLKFPWIGVVN 900
Query: 255 RSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGL 314
RSQ+DI N + A E E+F S P Y LA+ G LAK L+K L IK+ +PG+
Sbjct: 901 RSQQDINKNVDMIAARRREREYFNSTPEYKHLANRMGSEHLAKMLSKHLETVIKSKIPGI 1080
Query: 315 KARISTSLVALAKEHASYGEITESKAGQGAL--VLNILSKYCDAFSSMLEGKNKEMSTCE 372
++ I+ ++ L E G+ + AG G L ++ I + F L+G
Sbjct: 1081QSLINKTIAELEAELTRLGKPVAADAG-GKLYAIMEICRSFDQIFKDHLDGVR------- 1236
Query: 373 LSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFVRR 432
GG +I+ +F + +L+ + + L+ ++IR + A G + L PE + +
Sbjct: 1237-PGGDKIYNVFDNQLPAALKRLQFDKQLSMENIRKLITEADGYQPHLIAPEQGYRRLIES 1413
Query: 433 QISRLLDPSLQCARFIYDELMKISHRCM--FTELQRFPFLRKHMDEVIGNFLRDGLEPSE 490
+ + P+ ++ L + H+ M +L+++P LR + + L + S+
Sbjct: 1414SLITIRGPAESAVDAVHSLLKDLVHKAMSETLDLKQYPGLRVEVGAASVDSLERMRDESK 1593
Query: 491 TMITHIIEMEMDYI 504
+++ME Y+
Sbjct: 1594RATLQLVDMECGYL 1635
>TC209329 similar to UP|Q9FNX5 (Q9FNX5) Dynamin-like protein DLP2
(AT3g60190/T2O9_170), partial (58%)
Length = 1200
Score = 280 bits (715), Expect = 2e-75
Identities = 158/352 (44%), Positives = 217/352 (60%), Gaps = 10/352 (2%)
Frame = +3
Query: 22 SVISLVNRLQDIFSRVGNQSTIID---------LPQVAVVGSQSSGKSSVLESLVGRDFL 72
S+I LVNR+Q + +G+ + LP VAVVG QSSGKSSVLES+VGRDFL
Sbjct: 117 SLIGLVNRIQQACTVLGDYGAADNNAFSSLWEALPSVAVVGGQSSGKSSVLESIVGRDFL 296
Query: 73 PRGTDICTRRPLVLQLVHTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKG 132
PRG+ I TRRPLVLQL E+ EFLH+PG+K D+ +R EIQ ETDR G K
Sbjct: 297 PRGSGIVTRRPLVLQLHKVDGKAKEYAEFLHMPGQKITDYAIVRQEIQNETDRVTGRTKQ 476
Query: 133 VSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAV 192
+S I L I+SP+V+++TL+DLPG+TKV V QP I I M+ S++ P C+ILA+
Sbjct: 477 ISPVPIHLSIYSPHVVNLTLIDLPGLTKVAVEGQPETIAQDIENMVRSFVDKPNCIILAI 656
Query: 193 TPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGV 252
+PAN D+A SDA+++++ DP G RT GV+TKLD+MDRGT+A ++L G+ L+ +VGV
Sbjct: 657 SPANQDIATSDAIKLSREVDPSGERTFGVLTKLDLMDRGTNALDVLEGRSYRLQHPWVGV 836
Query: 253 VNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLP 312
VNRSQ DI N + A E E+F + P Y LA+ G LAK L++ L I+ +P
Sbjct: 837 VNRSQADINKNVDMIVARRKESEYFETSPDYGHLANKMGSVYLAKLLSQHLESVIRQRIP 1016
Query: 313 GLKARISTSLVALAKEHASYGEITESKAG-QGALVLNILSKYCDAFSSMLEG 363
+ + I+ ++ L E G + AG Q +L + + F L+G
Sbjct: 1017NITSLINKTIEELESEMNQIGRPIAADAGAQLYTILELCRAFDRVFKEHLDG 1172
>AW760191
Length = 420
Score = 255 bits (652), Expect = 5e-68
Identities = 130/138 (94%), Positives = 134/138 (96%)
Frame = +2
Query: 111 DFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDI 170
DF++IR EIQAETDREAG NKGVS+KQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDI
Sbjct: 5 DFSEIRREIQAETDREAGGNKGVSDKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDI 184
Query: 171 EARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDR 230
EARIRTMIMSYIK PTCLILAVTPANSDLANSDALQMA +ADPDGNRTIGVITKLDIMDR
Sbjct: 185 EARIRTMIMSYIKTPTCLILAVTPANSDLANSDALQMAGIADPDGNRTIGVITKLDIMDR 364
Query: 231 GTDARNLLLGKVIPLRLG 248
GTDARNLLLGKVIPLRLG
Sbjct: 365 GTDARNLLLGKVIPLRLG 418
>AW309416
Length = 532
Score = 251 bits (641), Expect = 9e-67
Identities = 130/171 (76%), Positives = 147/171 (85%), Gaps = 1/171 (0%)
Frame = +2
Query: 84 LVLQLVHTKP-SHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKI 142
LVLQLV TKP S EFGEFLHLPGRKFHDF+QIR EIQ ETDREAG NKGVS+KQIRLK+
Sbjct: 2 LVLQLVQTKPPSQDEFGEFLHLPGRKFHDFSQIRAEIQVETDREAGGNKGVSDKQIRLKM 181
Query: 143 FSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANS 202
PNVLDITLVDLP ITKVP+G+QPSDIEA+I TMIMSYIK PTC+IL VTPANSDL NS
Sbjct: 182 VFPNVLDITLVDLPDITKVPIGNQPSDIEAQIITMIMSYIKTPTCIILTVTPANSDLTNS 361
Query: 203 DALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVV 253
+ALQ+A +ADP+ NRTIG+ITK +I+DR T+ RNL L + IPLRL YV +V
Sbjct: 362 NALQIAGIADPNRNRTIGIITKCNIIDRSTNTRNL*LREXIPLRLEYVRMV 514
>TC220183 weakly similar to UP|Q8LPH8 (Q8LPH8) Dynamin-like protein, partial
(9%)
Length = 714
Score = 155 bits (393), Expect = 5e-38
Identities = 77/99 (77%), Positives = 89/99 (89%), Gaps = 1/99 (1%)
Frame = +3
Query: 676 IMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRELLRAYQQAFRDLDE 735
IM+FLV+ K ELHNVFI+KLYR++LFEEML+EP E+A KRKRCRELLRAYQQAF+DL+E
Sbjct: 3 IMYFLVDKAKGELHNVFIKKLYRDNLFEEMLREPDEIALKRKRCRELLRAYQQAFKDLEE 182
Query: 736 LPLEAETVERGYGSPEATGLPRIRGLPSSSMYS-SSLGD 773
LPLEA+TVERGYG PE TGLP+I GLP+SSMYS SS GD
Sbjct: 183 LPLEADTVERGYGLPEKTGLPKINGLPTSSMYSASSSGD 299
>BM521552 similar to PIR|T04982|T049 dynamin-like protein ADL2 - Arabidopsis
thaliana, partial (11%)
Length = 429
Score = 154 bits (390), Expect = 1e-37
Identities = 76/97 (78%), Positives = 87/97 (89%)
Frame = +2
Query: 635 VLRPSESNSEMEGVEITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIE 694
VLR SES SE E ++ITVTKLLLRSYYDI RKN+EDLVPKAIM+FLV+ K ELHNVFI+
Sbjct: 2 VLRSSESYSETEAIDITVTKLLLRSYYDIVRKNVEDLVPKAIMYFLVDKAKGELHNVFIK 181
Query: 695 KLYREDLFEEMLQEPAEVAAKRKRCRELLRAYQQAFR 731
KLYR++LFEEML+EP E+A KRKRCRELLRAYQQAF+
Sbjct: 182 KLYRDNLFEEMLREPDEIALKRKRCRELLRAYQQAFK 292
>AI900488
Length = 346
Score = 144 bits (363), Expect = 1e-34
Identities = 78/112 (69%), Positives = 85/112 (75%), Gaps = 2/112 (1%)
Frame = +3
Query: 499 MEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSL--DALESDKGLASERNGKSRGI 556
ME++YINTSH NFIGGSKALE A QQTK S VS PVS + LESDKG SER+ KSR I
Sbjct: 6 MELNYINTSHHNFIGGSKALEIASQQTKSSMVSIPVSRQKEVLESDKGSVSERSVKSRAI 185
Query: 557 IARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYVKEN 608
+ARQANGVV + GV ASD EKV SGN GGSSW ISSIFGG D+ VKEN
Sbjct: 186 LARQANGVVTEPGVHAASDAEKVVSSGNTGGSSWGISSIFGGGDSHTIVKEN 341
>AW101443
Length = 399
Score = 128 bits (321), Expect = 1e-29
Identities = 69/100 (69%), Positives = 80/100 (80%), Gaps = 5/100 (5%)
Frame = +3
Query: 26 LVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTRRPLV 85
++ RL D FSRV +QSTI DLPQV VVGS+SSGKSS+LE+ VGRDFLPRG DICT RPLV
Sbjct: 66 VIYRL*DFFSRVESQSTI-DLPQVTVVGSRSSGKSSILEAFVGRDFLPRGNDICTCRPLV 242
Query: 86 LQLVHTK-----PSHPEFGEFLHLPGRKFHDFTQIRHEIQ 120
LQLV TK ++ E+G FLHLP RKFHDF++IR EIQ
Sbjct: 243 LQLV*TKRKPNLDNNDEYGVFLHLPDRKFHDFSEIRREIQ 362
>CA800458
Length = 433
Score = 116 bits (290), Expect = 4e-26
Identities = 62/141 (43%), Positives = 93/141 (64%)
Frame = +1
Query: 182 IKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGK 241
++ P C+ILA++PAN D+A SDA+++A+ DP G RT GV+TKLD+MD+GT+A ++L G+
Sbjct: 1 VEKPNCIILAISPANQDIATSDAIKIAREVDPSGERTFGVVTKLDLMDKGTNAVDVLEGR 180
Query: 242 VIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNK 301
L+ +VG+VNRSQ DI N + A E E+F + P Y LA G LAK L++
Sbjct: 181 QYRLQHPWVGIVNRSQADINRNVDMIAARRKEREYFET-PEYGHLAHKMGSEYLAKLLSQ 357
Query: 302 ILAQHIKAVLPGLKARISTSL 322
L Q I+ +P + A I+ ++
Sbjct: 358 HLEQVIRQKIPSIIALINKTI 420
>TC231772 similar to UP|Q9FNX5 (Q9FNX5) Dynamin-like protein DLP2
(AT3g60190/T2O9_170), partial (47%)
Length = 932
Score = 104 bits (259), Expect = 2e-22
Identities = 85/288 (29%), Positives = 135/288 (46%), Gaps = 3/288 (1%)
Frame = +2
Query: 220 GVITKLDIMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRS 279
GV+TKLD+MD+GT+A ++L G+ L+ +VG+VNRSQ DI N + A E E+F +
Sbjct: 2 GVLTKLDLMDKGTNALDVLEGRSYRLQHPWVGIVNRSQADINRNVDMIVARRKEREYFAT 181
Query: 280 RPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESK 339
Y LA+ G LAK L++ L I+A +P + + I+ S+ L E G
Sbjct: 182 SSDYGHLANKMGSEYLAKLLSQHLESVIRARIPSITSLINKSIEELESEMDHLGRPIALD 361
Query: 340 AGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCED 399
A GA + IL + C AF + KE GG RI+ +F + +L +
Sbjct: 362 A--GAQLYTIL-ELCRAFERIF----KEHLDGGRPGGDRIYNVFDNQLPAALRKLPLDRH 520
Query: 400 LTDDDIRTALQNATGPKSALFIPEVPFEIFVRRQISRLLDP---SLQCARFIYDELMKIS 456
L+ ++R + A G + L PE + + + P S+ F+ EL++ S
Sbjct: 521 LSLQNVRKVVSEADGYQPHLIAPEQGYRRLIEGALGYFRGPAEASVDAVNFVLKELVRKS 700
Query: 457 HRCMFTELQRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYI 504
EL+RFP + + L E S+ +++ME Y+
Sbjct: 701 -IAETKELKRFPTFQAELAAAANEALERFREESKKTTVRLVDMESSYL 841
>AI443755 similar to GP|19032339|dbj dynamin like protein 2b {Arabidopsis
thaliana}, partial (7%)
Length = 319
Score = 85.5 bits (210), Expect = 8e-17
Identities = 47/56 (83%), Positives = 52/56 (91%)
Frame = +3
Query: 12 ASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLV 67
A++ PLGSSVISLVNRLQDIF+RVG+QST IDLPQVAVVGSQSSGKSSVLE+LV
Sbjct: 153 AASDTAPLGSSVISLVNRLQDIFARVGSQST-IDLPQVAVVGSQSSGKSSVLEALV 317
>BM884967
Length = 421
Score = 71.2 bits (173), Expect = 2e-12
Identities = 39/45 (86%), Positives = 41/45 (90%)
Frame = +3
Query: 11 AASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQ 55
AAS A PLGSSVISLVNRLQDIF+RVG+QST IDLPQVAVVGSQ
Sbjct: 288 AASAATAPLGSSVISLVNRLQDIFARVGSQST-IDLPQVAVVGSQ 419
>TC206342
Length = 724
Score = 66.6 bits (161), Expect = 4e-11
Identities = 36/47 (76%), Positives = 40/47 (84%), Gaps = 2/47 (4%)
Frame = +2
Query: 729 AFRDLDELPLEAETVERGYGSPE-ATGLPRIRGLPSSSMYS-SSLGD 773
A +DL+ELPLEAETVERGY PE TGLP+IRGLP+SSMYS SS GD
Sbjct: 8 AXKDLNELPLEAETVERGYSLPEITTGLPKIRGLPTSSMYSTSSSGD 148
>BQ094303 PIR|S63667|S63 phragmoplastin 12 - soybean, partial (13%)
Length = 423
Score = 61.6 bits (148), Expect = 1e-09
Identities = 37/78 (47%), Positives = 52/78 (66%), Gaps = 13/78 (16%)
Frame = +3
Query: 22 SVISLVNRLQDIFSRVGNQS------TIID-LPQVAVVGSQSSGKSSVLESLVGRDFLPR 74
++ISLVN++Q + +G+ T+ D LP +AVVG QSSGKSSVLES+VG+DFLPR
Sbjct: 105 NLISLVNKIQRACTALGDHGENSALPTLWDSLPAIAVVGGQSSGKSSVLESVVGKDFLPR 284
Query: 75 GT------DICTRRPLVL 86
G+ +C + LV+
Sbjct: 285 GSGRRRSLSVCLKSMLVI 338
>CD486929
Length = 229
Score = 53.5 bits (127), Expect = 3e-07
Identities = 25/30 (83%), Positives = 28/30 (93%)
Frame = +1
Query: 46 LPQVAVVGSQSSGKSSVLESLVGRDFLPRG 75
LP +AVVG QSSGKSSVLES+VG+DFLPRG
Sbjct: 70 LPAIAVVGGQSSGKSSVLESVVGKDFLPRG 159
>AW755897 homologue to PIR|S63667|S63 phragmoplastin 12 - soybean, partial
(23%)
Length = 438
Score = 52.8 bits (125), Expect = 6e-07
Identities = 40/148 (27%), Positives = 69/148 (46%), Gaps = 1/148 (0%)
Frame = +3
Query: 277 FRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGE-I 335
F + P Y LA G LAK L+K L IK+ +PG+++ IS ++ L E + G+ +
Sbjct: 3 FSNTPEYKHLAHRMGSEHLAKMLSKHLEAVIKSKIPGIQSLISKTIAELEAELSRLGKPV 182
Query: 336 TESKAGQGALVLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVD 395
G+ V+ I + F L+G GG +I+ +F + +L+ +
Sbjct: 183 AADDGGKLYAVMEICRSFDHIFKEHLDGVR--------PGGDKIYNVFDNQLPAALKRLQ 338
Query: 396 PCEDLTDDDIRTALQNATGPKSALFIPE 423
+ L+ ++IR + A G + L PE
Sbjct: 339 FDKQLSMENIRKLITEADGYQPHLIAPE 422
>TC207743 similar to UP|Q8S3C8 (Q8S3C8) Dynamin-like protein C, partial (20%)
Length = 663
Score = 40.8 bits (94), Expect = 0.002
Identities = 31/129 (24%), Positives = 57/129 (44%), Gaps = 2/129 (1%)
Frame = +1
Query: 610 STKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYYDIARKNIE 669
S K H +P N + PP + ++++ G + SY + ++
Sbjct: 16 SRKIHFDPEKNPNGPPNPNRNGPPNMDSYTDNHLRKIGSNVN-------SYIGMVCDTLK 174
Query: 670 DLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFE--EMLQEPAEVAAKRKRCRELLRAYQ 727
+ +PKA++H V KR L N F + R++ + ML E + +R + + L Y+
Sbjct: 175 NTIPKAVVHCQVREAKRSLLNHFYVHVGRKEKEKLGAMLDEDPALMDRRNQIAKRLELYK 354
Query: 728 QAFRDLDEL 736
QA D+D +
Sbjct: 355 QARDDIDSV 381
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,255,001
Number of Sequences: 63676
Number of extensions: 367100
Number of successful extensions: 1808
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1787
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1797
length of query: 773
length of database: 12,639,632
effective HSP length: 105
effective length of query: 668
effective length of database: 5,953,652
effective search space: 3977039536
effective search space used: 3977039536
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0100c.1


