

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100b.1
(354 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
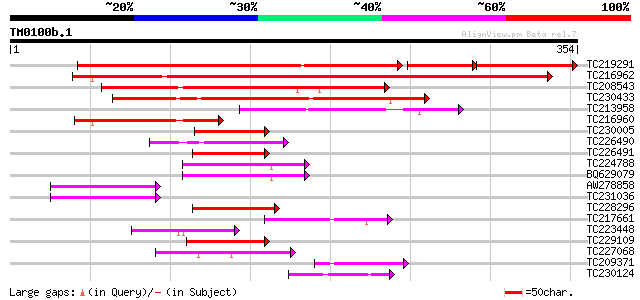
Score E
Sequences producing significant alignments: (bits) Value
TC219291 similar to UP|Q6J3Q5 (Q6J3Q5) Male-specific transcripti... 330 e-103
TC216962 similar to UP|Q6J3Q5 (Q6J3Q5) Male-specific transcripti... 259 1e-69
TC208543 similar to GB|BAD02931.1|38603660|AB119061 GATA-type zi... 189 2e-48
TC230433 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (45%) 174 4e-44
TC213958 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (29%) 109 2e-24
TC216960 similar to GB|BAD02931.1|38603660|AB119061 GATA-type zi... 85 4e-17
TC230005 similar to UP|Q6UEI7 (Q6UEI7) Timing of CAB expression ... 60 1e-09
TC226490 similar to UP|O81035 (O81035) Expressed protein (Pseudo... 59 3e-09
TC226491 homologue to UP|O81035 (O81035) Expressed protein (Pseu... 57 1e-08
TC224788 weakly similar to UP|Q6UV11 (Q6UV11) Pseudo-response re... 55 4e-08
BQ629079 similar to GP|10281004|dbj pseudo-response regulator 7 ... 55 5e-08
AW278858 54 1e-07
TC231036 similar to UP|Q9LU42 (Q9LU42) Emb|CAB45082.1 (GATA-type... 54 1e-07
TC228296 similar to UP|O81035 (O81035) Expressed protein (Pseudo... 54 1e-07
TC217661 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (47%) 54 1e-07
TC223448 similar to UP|Q6J3Q5 (Q6J3Q5) Male-specific transcripti... 53 2e-07
TC229109 similar to UP|Q6LA42 (Q6LA42) Pseudo-response regulator... 51 7e-07
TC227068 similar to UP|Q6UV11 (Q6UV11) Pseudo-response regulator... 50 1e-06
TC209371 similar to PIR|T52104|T52104 GATA-binding transcription... 49 3e-06
TC230124 similar to UP|Q76DY1 (Q76DY1) AG-motif binding protein-... 49 3e-06
>TC219291 similar to UP|Q6J3Q5 (Q6J3Q5) Male-specific transcription factor
M88B7.2, partial (29%)
Length = 1458
Score = 330 bits (847), Expect(2) = e-103
Identities = 165/204 (80%), Positives = 179/204 (86%), Gaps = 1/204 (0%)
Frame = +3
Query: 43 AASASNAS-APHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQ 101
A +NAS A H RA ELTISFEGEVYVFPAVTPEKVQAVLLLLG +++ NS P S +L
Sbjct: 9 AVPVTNASSAMHARASELTISFEGEVYVFPAVTPEKVQAVLLLLGAQEMTNSAPTSDILL 188
Query: 102 QKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQ 161
Q+N+ I+ INDPS+SS LSRRFASLVRFREKRKERCFEKKIRY+CRKEVAQRMHRKNGQ
Sbjct: 189 QQNYQDIREINDPSRSSKLSRRFASLVRFREKRKERCFEKKIRYSCRKEVAQRMHRKNGQ 368
Query: 162 FASLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACG 221
FAS+KEDYKSPA N DSSNGT PCP+S ERRCQHCG SEKSTPAMRRGPAGPRSLCNACG
Sbjct: 369 FASMKEDYKSPAENWDSSNGT-PCPDSTERRCQHCGISEKSTPAMRRGPAGPRSLCNACG 545
Query: 222 LMWANKGTLRDLSKAAKTPYEQNE 245
LMWANKGTLRDLSKA + +EQNE
Sbjct: 546 LMWANKGTLRDLSKAGRIAFEQNE 617
Score = 100 bits (249), Expect = 1e-21
Identities = 49/64 (76%), Positives = 58/64 (90%), Gaps = 1/64 (1%)
Frame = +2
Query: 292 QDILGTAQTVTDNLSIQLENHALSLDEQD-IEDLADASGTEFGIPAGFDDQIDIDVSNMR 350
Q I+GTA++VT+NLS++LENHAL L EQD +EDLADASGTEF IPAGFDDQ+DID +NMR
Sbjct: 1019 QFIIGTAESVTNNLSVRLENHALILHEQDTLEDLADASGTEFEIPAGFDDQVDIDDANMR 1198
Query: 351 TYWL 354
TYWL
Sbjct: 1199 TYWL 1210
Score = 63.2 bits (152), Expect(2) = e-103
Identities = 29/44 (65%), Positives = 36/44 (80%)
Frame = +2
Query: 249 SADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRETPEKKNEQ 292
+AD+KPSTTE ++S A Q +EG+PEE KPV MDSR +PEK NEQ
Sbjct: 617 TADIKPSTTEAKHSYAKQGKEGSPEETKPVQMDSRRSPEKTNEQ 748
>TC216962 similar to UP|Q6J3Q5 (Q6J3Q5) Male-specific transcription factor
M88B7.2, partial (31%)
Length = 1446
Score = 259 bits (662), Expect = 1e-69
Identities = 152/307 (49%), Positives = 199/307 (64%), Gaps = 7/307 (2%)
Frame = +3
Query: 40 ATVAASASNAS--APHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNS 97
A AAS ++ + A R ELT+SFEGEVYVFPA+TP+KVQAVLLLLG +D+ P
Sbjct: 300 ANAAASVNHEAVVAMPSRTSELTLSFEGEVYVFPAITPQKVQAVLLLLGGRDVQARVP-- 473
Query: 98 GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHR 157
++Q +G+ D + SNLSRR ASLVRFREKRKERCF+KKIRY+ RKEVAQRMHR
Sbjct: 474 -AVEQPFDQSNRGMGDTPKRSNLSRRIASLVRFREKRKERCFDKKIRYSVRKEVAQRMHR 650
Query: 158 KNGQFASLKEDYKSPAGNLDSSNGTPPCPESIE-RRCQHCGTSEKSTPAMRRGPAGPRSL 216
KNGQFASLKE S + S+G S RRC HCG E +TPAMRRGPAGPR+L
Sbjct: 651 KNGQFASLKESPGSSNWDSAQSSGQVGTSHSESVRRCHHCGVGENNTPAMRRGPAGPRTL 830
Query: 217 CNACGLMWANKGTLRDLSKAAKT-PYEQNERDTSADMKPSTTEPENSCADQAEEGTPEE- 274
CNACGLMWANKGTLRDLSK + EQ++ DT D+KP++ E+G+ E+
Sbjct: 831 CNACGLMWANKGTLRDLSKGGRNLSVEQSDLDTPIDVKPTSVLEGELPGIHDEQGSSEDP 1010
Query: 275 NKPVPMD-SRETPEKKNEQDILGTAQTVTDNLSIQLENHALSLDEQD-IEDLADASGTEF 332
+K D S +++++ TA+ T+ L + + + + + EQ+ + +L++ S T+
Sbjct: 1011SKSNAADGSSNHAVNPSDEELPETAEHFTNVLPLGIGHSSTNDSEQEPLVELSNPSDTDI 1190
Query: 333 GIPAGFD 339
IP FD
Sbjct: 1191DIPGNFD 1211
>TC208543 similar to GB|BAD02931.1|38603660|AB119061 GATA-type zinc finger
protein {Arabidopsis thaliana;} , partial (50%)
Length = 929
Score = 189 bits (480), Expect = 2e-48
Identities = 103/186 (55%), Positives = 127/186 (67%), Gaps = 6/186 (3%)
Frame = +2
Query: 58 ELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGINDPSQS 117
+LT+SF+G+VYVF +V+PEKVQAVLLLLG ++IP + P + N +G Q
Sbjct: 26 QLTLSFQGQVYVFDSVSPEKVQAVLLLLGGREIPPTMPAMPVSPNHNN---RGYTGTPQK 196
Query: 118 SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLD 177
++ +R ASL+RFREKRKER ++KKIRYT RKEVA RM R GQF S K + A N
Sbjct: 197 FSVPQRLASLIRFREKRKERNYDKKIRYTVRKEVALRMQRNKGQFTSSKSNNDESASNAT 376
Query: 178 S----SNGTPPCPESIERR--CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLR 231
+ N T S ++ C+HCG SEKSTP MRRGP GPR+LCNACGLMWANKG LR
Sbjct: 377 NWGMDENWTADNSGSQQQDIVCRHCGISEKSTPMMRRGPEGPRTLCNACGLMWANKGILR 556
Query: 232 DLSKAA 237
DLS+AA
Sbjct: 557 DLSRAA 574
>TC230433 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (45%)
Length = 889
Score = 174 bits (442), Expect = 4e-44
Identities = 101/203 (49%), Positives = 127/203 (61%), Gaps = 5/203 (2%)
Frame = +1
Query: 65 GEVYVFPAVTPEKVQAVLLLLGQKDIPNS-TPNSGLLQQKNFPQIKGINDPSQSSNLSRR 123
G+VYVF AVTP+KVQAVLLLLG ++ + +P Q N Q + P S L R
Sbjct: 1 GQVYVFDAVTPDKVQAVLLLLGGCELSSGGSPCVDPGAQHN--QRGSMEFPKCS--LPHR 168
Query: 124 FASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTP 183
ASL RFR+KRKERCF+KK+RY+ R+EVA RMHR GQF S K+ + + D +G
Sbjct: 169 AASLHRFRQKRKERCFDKKVRYSVRQEVALRMHRNKGQFTSSKKQDGANSYGTDQDSGQD 348
Query: 184 PCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKA----AKT 239
E C HCG S KSTP MRRGP+GPRSLCNACGL WAN+G LRDLSK +
Sbjct: 349 DSQS--ETSCTHCGISSKSTPMMRRGPSGPRSLCNACGLFWANRGALRDLSKRNQEHSLP 522
Query: 240 PYEQNERDTSADMKPSTTEPENS 262
P EQ + +D + +T +P ++
Sbjct: 523 PVEQVDEGNDSDCRTATADPAHN 591
>TC213958 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (29%)
Length = 659
Score = 109 bits (273), Expect = 2e-24
Identities = 63/142 (44%), Positives = 78/142 (54%), Gaps = 2/142 (1%)
Frame = +2
Query: 144 RYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKST 203
RY+ R+EVA RMHR GQF S K + + D +G +S C HCG S KST
Sbjct: 2 RYSVRQEVALRMHRNKGQFTSSKNQDGTNSWGSDQESGQDAV-QSETLCCTHCGISSKST 178
Query: 204 PAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADMKP--STTEPEN 261
P MRRGP+GPRSLCNACGL WAN+GTLRDLSK R+ + P E N
Sbjct: 179 PMMRRGPSGPRSLCNACGLFWANRGTLRDLSK----------RNLEHSLAPPEQVDEGSN 328
Query: 262 SCADQAEEGTPEENKPVPMDSR 283
+ A G P ++ + DS+
Sbjct: 329 NNAFDFRSGIPAQHNNLVNDSK 394
>TC216960 similar to GB|BAD02931.1|38603660|AB119061 GATA-type zinc finger
protein {Arabidopsis thaliana;} , partial (14%)
Length = 585
Score = 85.1 bits (209), Expect = 4e-17
Identities = 53/96 (55%), Positives = 63/96 (65%), Gaps = 3/96 (3%)
Frame = +3
Query: 41 TVAASASNAS--APHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPN-S 97
T AAS ++ + A R ELT+SFEGEVYVFPAVTP+KVQAVLLLLG +D+ P
Sbjct: 309 TNAASVNHEAVVAMPSRTSELTLSFEGEVYVFPAVTPQKVQAVLLLLGGRDVQAGVPAVE 488
Query: 98 GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREK 133
Q N + + D + SNLSRR ASLVRFREK
Sbjct: 489 PPFDQSN----RDMGDTPKRSNLSRRIASLVRFREK 584
>TC230005 similar to UP|Q6UEI7 (Q6UEI7) Timing of CAB expression 1, partial
(15%)
Length = 856
Score = 60.1 bits (144), Expect = 1e-09
Identities = 28/47 (59%), Positives = 36/47 (76%)
Frame = +3
Query: 116 QSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
Q S + RR A+L++FR+KRKERCF+KKIRY RK +A+R R GQF
Sbjct: 99 QISKVDRREAALMKFRQKRKERCFDKKIRYVNRKRLAERRPRVRGQF 239
>TC226490 similar to UP|O81035 (O81035) Expressed protein (Pseudo-response
regulator 9) (Timing of CAB expression 1-like protein),
partial (9%)
Length = 1411
Score = 59.3 bits (142), Expect = 3e-09
Identities = 36/87 (41%), Positives = 49/87 (55%)
Frame = +1
Query: 88 KDIPNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTC 147
K+ P+ +SG F + DP +SS +R A LV+FR KRKERCFEKK+RY
Sbjct: 862 KNAPDGFSDSGCHNYDGFR----VTDPHRSS---QREAVLVKFRLKRKERCFEKKVRYQS 1020
Query: 148 RKEVAQRMHRKNGQFASLKEDYKSPAG 174
RK +A++ R GQF + + AG
Sbjct: 1021RKRLAEQRPRVKGQFVRQHDHPVAEAG 1101
>TC226491 homologue to UP|O81035 (O81035) Expressed protein (Pseudo-response
regulator 9) (Timing of CAB expression 1-like protein),
partial (9%)
Length = 730
Score = 57.0 bits (136), Expect = 1e-08
Identities = 27/48 (56%), Positives = 36/48 (74%)
Frame = +3
Query: 115 SQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
+ S + S+R A+LV+FR KRKERCFEKK+RY RK +A++ R GQF
Sbjct: 117 TDSHHSSQREAALVKFRLKRKERCFEKKVRYQSRKRLAEQRPRVKGQF 260
>TC224788 weakly similar to UP|Q6UV11 (Q6UV11) Pseudo-response regulator
protein (Fragment), partial (12%)
Length = 1699
Score = 55.5 bits (132), Expect = 4e-08
Identities = 31/84 (36%), Positives = 47/84 (55%), Gaps = 5/84 (5%)
Frame = +3
Query: 109 KGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF-----A 163
K + + L+ R A+L +FR KRKERCFEKK+RY RK++A++ R GQF +
Sbjct: 660 KSFGNGADEGRLALREAALTKFRLKRKERCFEKKVRYHSRKKLAEQRPRIRGQFVRRIVS 839
Query: 164 SLKEDYKSPAGNLDSSNGTPPCPE 187
KE+ + NL + + P+
Sbjct: 840 EGKEEKDKQSDNLVPGDNSIDIPQ 911
>BQ629079 similar to GP|10281004|dbj pseudo-response regulator 7 {Arabidopsis
thaliana}, partial (7%)
Length = 436
Score = 55.1 bits (131), Expect = 5e-08
Identities = 31/84 (36%), Positives = 48/84 (56%), Gaps = 5/84 (5%)
Frame = +3
Query: 109 KGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF-----A 163
K I + + L+ R A+L +FR KRKERCFEK++RY RK++A++ R GQF +
Sbjct: 129 KSIGNGTDEVRLALREAALTKFRLKRKERCFEKRVRYHSRKKLAEQRPRIKGQFVRRIVS 308
Query: 164 SLKEDYKSPAGNLDSSNGTPPCPE 187
KE+ + NL + + P+
Sbjct: 309 EGKEEKDKQSDNLVPGDNSIDIPQ 380
>AW278858
Length = 430
Score = 53.5 bits (127), Expect = 1e-07
Identities = 26/69 (37%), Positives = 41/69 (58%)
Frame = +2
Query: 26 VHFANGADEGQHAPATVAASASNASAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLL 85
+H+++ E A ++ + +LT+SF G+VYVF AVTP+KVQAVLLLL
Sbjct: 224 IHYSSHTIEDDGAAVEDVSAVPGPEISIDNSSQLTLSFRGQVYVFDAVTPDKVQAVLLLL 403
Query: 86 GQKDIPNST 94
G ++ + +
Sbjct: 404 GGNELTSGS 430
>TC231036 similar to UP|Q9LU42 (Q9LU42) Emb|CAB45082.1 (GATA-type zinc finger
protein), partial (12%)
Length = 637
Score = 53.5 bits (127), Expect = 1e-07
Identities = 26/69 (37%), Positives = 41/69 (58%)
Frame = +3
Query: 26 VHFANGADEGQHAPATVAASASNASAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLL 85
+H+++ E A ++ + +LT+SF G+VYVF AVTP+KVQAVLLLL
Sbjct: 345 IHYSSHTIEDDGAAVEDVSAVPGPEISIDNSSQLTLSFRGQVYVFDAVTPDKVQAVLLLL 524
Query: 86 GQKDIPNST 94
G ++ + +
Sbjct: 525 GGNELTSGS 551
>TC228296 similar to UP|O81035 (O81035) Expressed protein (Pseudo-response
regulator 9) (Timing of CAB expression 1-like protein),
partial (14%)
Length = 1371
Score = 53.5 bits (127), Expect = 1e-07
Identities = 25/54 (46%), Positives = 35/54 (64%)
Frame = +3
Query: 115 SQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKED 168
+ S S+R A+L +FR KRK+RC+EKK+RY RK +A++ R GQF D
Sbjct: 828 TDSHRTSQREAALTKFRLKRKDRCYEKKVRYQSRKRLAEQRPRVKGQFVRQVHD 989
>TC217661 similar to UP|Q9FJ10 (Q9FJ10) Gb|AAF63824.1, partial (47%)
Length = 765
Score = 53.5 bits (127), Expect = 1e-07
Identities = 30/89 (33%), Positives = 46/89 (50%), Gaps = 9/89 (10%)
Frame = +2
Query: 160 GQFASLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNA 219
G+ + ++ + + N SS +P ++ C CGT++ TP R GPAGP+SLCNA
Sbjct: 191 GKGSEIEVEDSNSNPNAPSSGNSPSSNNEQKKTCADCGTTK--TPLWRGGPAGPKSLCNA 364
Query: 220 CG---------LMWANKGTLRDLSKAAKT 239
CG ++ NKG+ D K +T
Sbjct: 365 CGIRSRKKKRAILGINKGSNEDGRKGKRT 451
>TC223448 similar to UP|Q6J3Q5 (Q6J3Q5) Male-specific transcription factor
M88B7.2, partial (8%)
Length = 645
Score = 52.8 bits (125), Expect = 2e-07
Identities = 41/99 (41%), Positives = 47/99 (47%), Gaps = 32/99 (32%)
Frame = +3
Query: 77 KVQAVLLLLGQKDIPNSTPNSGLLQQKN----------------FPQ------------- 107
+VQAVLLLLG +++ NS P K F Q
Sbjct: 144 QVQAVLLLLGAQEMTNSAPT*LFFAAKLS*HYSISLGIAFLFDIFYQ*LLF*HIIKCCKT 323
Query: 108 ---IKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKI 143
+ G QSS +SRRFASLVRF EKRKERCFEKKI
Sbjct: 324 CYCMPGNK*SFQSSKISRRFASLVRFLEKRKERCFEKKI 440
>TC229109 similar to UP|Q6LA42 (Q6LA42) Pseudo-response regulator 5, partial
(7%)
Length = 1011
Score = 51.2 bits (121), Expect = 7e-07
Identities = 24/52 (46%), Positives = 36/52 (69%)
Frame = +2
Query: 111 INDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
+ + + S +R A+L +FR KRKERC+EKK+RY RK++A++ R GQF
Sbjct: 548 LTNNANSHRSIQREAALNKFRLKRKERCYEKKVRYESRKKLAEQRPRVKGQF 703
>TC227068 similar to UP|Q6UV11 (Q6UV11) Pseudo-response regulator protein
(Fragment), partial (26%)
Length = 1737
Score = 50.4 bits (119), Expect = 1e-06
Identities = 30/92 (32%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Frame = +1
Query: 92 NSTPNSGLLQQKNFPQIKGINDPSQ--SSNLSRRFASLVRFREKRKER---CFEKKIRYT 146
N+ N+GL G ++ + S+R +L +FR+KRKER CF KK+RY
Sbjct: 1192 NTESNNGLTGNSGSGDASGSGSANRVDQNKTSQREVALTKFRQKRKERRERCFHKKVRYQ 1371
Query: 147 CRKEVAQRMHRKNGQFASLKEDYKSPAGNLDS 178
RK +A++ R GQF + + DS
Sbjct: 1372 SRKRLAEQRPRFRGQFVRQTSNENASEATTDS 1467
>TC209371 similar to PIR|T52104|T52104 GATA-binding transcription factor
homolog 2 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (40%)
Length = 857
Score = 49.3 bits (116), Expect = 3e-06
Identities = 22/59 (37%), Positives = 32/59 (53%)
Frame = +2
Query: 191 RRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTS 249
RRC HC T + TP R GP GP++LCNACG+ + + + + AA + + S
Sbjct: 407 RRCSHCATDK--TPQWRTGPLGPKTLCNACGVRFKSGRLVPEYRPAASPTFVMTQHSNS 577
>TC230124 similar to UP|Q76DY1 (Q76DY1) AG-motif binding protein-3, partial
(26%)
Length = 894
Score = 48.9 bits (115), Expect = 3e-06
Identities = 26/66 (39%), Positives = 35/66 (52%)
Frame = +1
Query: 175 NLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLS 234
N ++N I RRC HC + TP R GP GP++LCNACG+ + G L
Sbjct: 142 NNTNNNNNDNVQHPIPRRCTHC--LAQRTPQWRAGPLGPKTLCNACGVRY-KSGRLLPEY 312
Query: 235 KAAKTP 240
+ AK+P
Sbjct: 313 RPAKSP 330
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,129,254
Number of Sequences: 63676
Number of extensions: 205392
Number of successful extensions: 1118
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 1077
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1101
length of query: 354
length of database: 12,639,632
effective HSP length: 98
effective length of query: 256
effective length of database: 6,399,384
effective search space: 1638242304
effective search space used: 1638242304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0100b.1


