

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100a.4
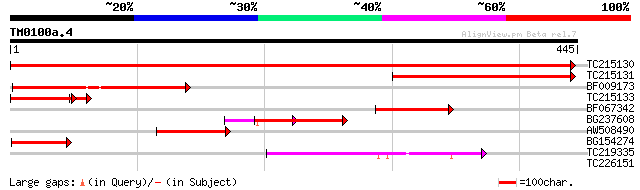
(445 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC215130 homologue to UP|O65744 (O65744) GDP dissociation inhibi... 766 0.0
TC215131 homologue to UP|O65744 (O65744) GDP dissociation inhibi... 243 1e-64
BF009173 similar to GP|3175990|emb GDP dissociation inhibitor {C... 143 2e-34
TC215133 homologue to UP|O65744 (O65744) GDP dissociation inhibi... 100 4e-25
BF067342 weakly similar to PIR|T01782|T017 GDP dissociation inhi... 84 1e-16
BG237608 homologue to PIR|T01782|T017 GDP dissociation inhibitor... 84 1e-16
AW508490 similar to GP|3175990|emb| GDP dissociation inhibitor {... 82 4e-16
BG154274 similar to PIR|T01782|T017 GDP dissociation inhibitor -... 74 2e-13
TC219335 weakly similar to UP|Q9LJ18 (Q9LJ18) ESTs AU056916(S209... 63 3e-10
TC226151 29 3.7
>TC215130 homologue to UP|O65744 (O65744) GDP dissociation inhibitor,
complete
Length = 1747
Score = 766 bits (1977), Expect = 0.0
Identities = 368/444 (82%), Positives = 405/444 (90%)
Frame = +3
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSLNLTQLWKRFRGDDK 60
MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGG STSLNL QLWKRFRGDDK
Sbjct: 123 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGESTSLNLIQLWKRFRGDDK 302
Query: 61 SPEHLGSSREFNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPA 120
P HLG+SR++N+DM PKF+MANG LVRVLIHTDVTKYL+FKAVDGSFV+NKGK++KVPA
Sbjct: 303 PPPHLGASRDYNIDMNPKFIMANGNLVRVLIHTDVTKYLSFKAVDGSFVFNKGKVHKVPA 482
Query: 121 TDVEALKSPLMGLFEKRRARKFFIYVQDYEVSDPKSHEGLDLNQVTARQLISKYGLEDDT 180
D+EALKSPLMGLFEKRRARKFFIYVQ+Y SDPK+HEG+DL +VT ++LI+KYGL+D+T
Sbjct: 483 NDMEALKSPLMGLFEKRRARKFFIYVQNYNESDPKTHEGMDLTRVTTKELIAKYGLDDNT 662
Query: 181 IDFIGHALALHLDDSYLDTPAKDFVERIKVYAESLARFQGGSPYIYPLYGLGELPQAFAR 240
IDFIGHALALH DD YL PA D V+R+K+YAESLARFQGGSPYIYPLYGLGELPQAFAR
Sbjct: 663 IDFIGHALALHSDDRYLAEPALDTVKRMKLYAESLARFQGGSPYIYPLYGLGELPQAFAR 842
Query: 241 LSAVYGGTYMLNKPECKVEFDSAGKAIGVTSEGETAKCKKVVCDPSYLPDKVRNVGKVAR 300
LSAVYGGTYMLNKPECKVEFD GK +GVTSEGETAKCKKVVCDPSYLP KVR VG+VAR
Sbjct: 843 LSAVYGGTYMLNKPECKVEFDEEGKVVGVTSEGETAKCKKVVCDPSYLPGKVRKVGRVAR 1022
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
AI IMSHPIP T+DSHS Q+ILPQKQLGRKSDMYLFCCSY HNVAPKGK+IAFV+TEAET
Sbjct: 1023AIAIMSHPIPSTDDSHSVQIILPQKQLGRKSDMYLFCCSYTHNVAPKGKFIAFVSTEAET 1202
Query: 361 DQPEVELKPGIDLLGPVDETFYDIYDRYEPTNDHAADGCFISKSYDATTHFETTVKDVVE 420
DQP +ELKPGIDLLGPVDE F+DIYDRYEP N+ D CF+S SYDATTHFE+TV DV+
Sbjct: 1203DQPAIELKPGIDLLGPVDEIFFDIYDRYEPVNEPTLDNCFVSTSYDATTHFESTVDDVLN 1382
Query: 421 IYSKITGKVLDLSVDLSAASATAE 444
+Y+ ITGKV+DLSVDLSAASA E
Sbjct: 1383MYTLITGKVIDLSVDLSAASAAEE 1454
>TC215131 homologue to UP|O65744 (O65744) GDP dissociation inhibitor, partial
(32%)
Length = 798
Score = 243 bits (621), Expect = 1e-64
Identities = 115/144 (79%), Positives = 127/144 (87%)
Frame = +1
Query: 301 AICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPKGKYIAFVTTEAET 360
AI IMSHPIP T+DSHS Q+ILPQKQLGRKSDMYLFCCSY HNVAPKGK+IAFV+TEAET
Sbjct: 1 AIAIMSHPIPSTDDSHSVQIILPQKQLGRKSDMYLFCCSYTHNVAPKGKFIAFVSTEAET 180
Query: 361 DQPEVELKPGIDLLGPVDETFYDIYDRYEPTNDHAADGCFISKSYDATTHFETTVKDVVE 420
DQP +ELKPGIDLLGPVDE F+DIYDRYEP N+ D CFIS SYDATTHFE+TV DV+
Sbjct: 181 DQPAIELKPGIDLLGPVDEIFFDIYDRYEPVNEPTLDNCFISTSYDATTHFESTVDDVLN 360
Query: 421 IYSKITGKVLDLSVDLSAASATAE 444
+Y+ ITGKV+DLSVDLSAASA E
Sbjct: 361 MYTLITGKVIDLSVDLSAASAAEE 432
>BF009173 similar to GP|3175990|emb GDP dissociation inhibitor {Cicer
arietinum}, partial (25%)
Length = 418
Score = 143 bits (360), Expect = 2e-34
Identities = 81/140 (57%), Positives = 95/140 (67%)
Frame = +2
Query: 3 EEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSLNLTQLWKRFRGDDKSP 62
EEY VIVLGT L ECIL+GLL V GL VLHMDRNDY GG STSLNL QLW+ FRGDDK P
Sbjct: 5 EEYYVIVLGTALTECILTGLLCVHGLIVLHMDRNDYLGGESTSLNLIQLWRTFRGDDK-P 181
Query: 63 EHLGSSREFNVDMIPKFMMANGALVRVLIHTDVTKYLNFKAVDGSFVYNKGKIYKVPATD 122
+ R+ + ++ VRVLIHTDVT YL K V GS V++ K++KVPA D
Sbjct: 182 APIRRCRD-STSLLTLSYPG*WQFVRVLIHTDVTMYLYIKDVHGSSVFDIRKVHKVPAHD 358
Query: 123 VEALKSPLMGLFEKRRARKF 142
+ AL PLM L E+RR K+
Sbjct: 359 M*ALMCPLMALLERRRPLKY 418
>TC215133 homologue to UP|O65744 (O65744) GDP dissociation inhibitor, partial
(14%)
Length = 418
Score = 99.8 bits (247), Expect(2) = 4e-25
Identities = 48/52 (92%), Positives = 50/52 (95%)
Frame = +2
Query: 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSLNLTQLW 52
MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGG STSLNL Q++
Sbjct: 122 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGESTSLNLIQVF 277
Score = 33.1 bits (74), Expect(2) = 4e-25
Identities = 13/17 (76%), Positives = 13/17 (76%)
Frame = +3
Query: 48 LTQLWKRFRGDDKSPEH 64
L QLWKRF GDDK P H
Sbjct: 366 LLQLWKRFXGDDKPPPH 416
>BF067342 weakly similar to PIR|T01782|T017 GDP dissociation inhibitor -
common tobacco, partial (13%)
Length = 184
Score = 84.3 bits (207), Expect = 1e-16
Identities = 38/61 (62%), Positives = 48/61 (78%)
Frame = +2
Query: 288 LPDKVRNVGKVARAICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVAPK 347
LP +VR VG+V+ AI IMSHPIP ++D+HS Q+I+P Q+G KS+MYLFCCSY H A K
Sbjct: 2 LPRRVRKVGRVSIAIPIMSHPIPTSDDTHSVQIIIPHIQMGLKSNMYLFCCSYTHIAATK 181
Query: 348 G 348
G
Sbjct: 182 G 184
>BG237608 homologue to PIR|T01782|T017 GDP dissociation inhibitor - common
tobacco, partial (13%)
Length = 432
Score = 84.0 bits (206), Expect = 1e-16
Identities = 48/76 (63%), Positives = 54/76 (70%), Gaps = 3/76 (3%)
Frame = +3
Query: 193 DDSYLDTPAK-DFVERIKVYAESLAR-FQGGSPYIY-PLYGLGELPQAFARLSAVYGGTY 249
D YL P+ V++ +V ++SL FQG Y LYGLGELPQAFARLSAVYGGTY
Sbjct: 78 DAXYLAEPSTWTTVKKNEVISQSLLHAFQGXVTLTYIALYGLGELPQAFARLSAVYGGTY 257
Query: 250 MLNKPECKVEFDSAGK 265
MLNKPECKVEFD GK
Sbjct: 258 MLNKPECKVEFDEEGK 305
Score = 47.4 bits (111), Expect = 1e-05
Identities = 32/66 (48%), Positives = 39/66 (58%), Gaps = 8/66 (12%)
Frame = +2
Query: 169 QLISKYGLEDDTIDFIGHALALHLD-------DSYLDTPAKDFVERIKVYAESLARF-QG 220
+LI+KYGL+D+TIDFIGHALALH ++D D E AESLAR +
Sbjct: 2 ELIAKYGLDDNTIDFIGHALALHSR*CAXLG*AEHVD----DREEE*SYIAESLARLSRX 169
Query: 221 GSPYIY 226
G YIY
Sbjct: 170 GHAYIY 187
>AW508490 similar to GP|3175990|emb| GDP dissociation inhibitor {Cicer
arietinum}, partial (12%)
Length = 190
Score = 82.4 bits (202), Expect = 4e-16
Identities = 38/58 (65%), Positives = 49/58 (83%)
Frame = +2
Query: 116 YKVPATDVEALKSPLMGLFEKRRARKFFIYVQDYEVSDPKSHEGLDLNQVTARQLISK 173
+KVPA ++ALKSPLMGLFEKRRARK FIYV +Y+ SD +HEG+DL +VT ++L+SK
Sbjct: 17 HKVPANYMKALKSPLMGLFEKRRARKLFIYVHNYDESDLSTHEGMDLTRVTTKELVSK 190
>BG154274 similar to PIR|T01782|T017 GDP dissociation inhibitor - common
tobacco, partial (10%)
Length = 142
Score = 73.6 bits (179), Expect = 2e-13
Identities = 37/47 (78%), Positives = 39/47 (82%)
Frame = +2
Query: 2 DEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSLNL 48
DEEYDVIVLGTGL EC+LSGLLS DGLKVLHMD D+Y G T LNL
Sbjct: 2 DEEYDVIVLGTGLNECMLSGLLSSDGLKVLHMD*TDFY*GEYT*LNL 142
>TC219335 weakly similar to UP|Q9LJ18 (Q9LJ18) ESTs AU056916(S20978), partial
(25%)
Length = 1389
Score = 62.8 bits (151), Expect = 3e-10
Identities = 55/198 (27%), Positives = 91/198 (45%), Gaps = 25/198 (12%)
Frame = +3
Query: 202 KDFVERIKVYAESLARFQGG-SPYIYPLYGLGELPQAFARLSAVYGGTYMLNKPECKVEF 260
KD ++ + +Y+ S+ RF +YP+YG GELPQAF R +AV G Y+L +
Sbjct: 21 KDGIDSLALYSSSVGRFPNAPGALLYPIYGEGELPQAFCRRAAVKGCIYVLRMSVVSLLM 200
Query: 261 DS-AGKAIGV-TSEGETAKCKKVVCDPSYL-------------PDKVRNV------GKVA 299
D G GV + G+ +++ DPS++ +K++ + G VA
Sbjct: 201 DKVTGSYKGVRLASGQDLYSHQLILDPSFMIPSPLSSSPRDFPSEKLQMLSQRDIKGMVA 380
Query: 300 RAICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHNVA--PKGKYIAFVTTE 357
R ICI I D S V+ P + L + + N+A P G +I +++T
Sbjct: 381 RGICITRSSIKP--DVSSCSVVYPPRSLYPEQVTSIKALQIGSNLAVCPAGTFILYLSTM 554
Query: 358 A-ETDQPEVELKPGIDLL 374
+ D+ + LK ++ L
Sbjct: 555 CNDADEGKKLLKAAMNAL 608
>TC226151
Length = 864
Score = 29.3 bits (64), Expect = 3.7
Identities = 25/94 (26%), Positives = 42/94 (44%), Gaps = 10/94 (10%)
Frame = +2
Query: 117 KVPATDVEA-----LKSPLMGLFEKRRARKFFIYVQ-----DYEVSDPKSHEGLDLNQVT 166
K ATDV LK + L +++ + K +Q DYEV + +
Sbjct: 341 KAEATDVLQKENLMLKEQIEALIKEKNSFKNAFRIQHERSADYEVKNQELQH-------- 496
Query: 167 ARQLISKYGLEDDTIDFIGHALALHLDDSYLDTP 200
+QL+S+Y + T++ +ALA+HL + P
Sbjct: 497 LKQLVSQYQEQIKTLEVNNYALAMHLKQAQQSNP 598
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,268,888
Number of Sequences: 63676
Number of extensions: 222430
Number of successful extensions: 834
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 832
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 833
length of query: 445
length of database: 12,639,632
effective HSP length: 100
effective length of query: 345
effective length of database: 6,272,032
effective search space: 2163851040
effective search space used: 2163851040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0100a.4


