

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098e.6
(402 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
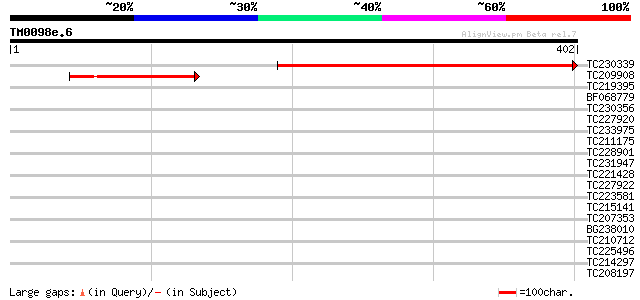
TC230339 homologue to UP|Q40093 (Q40093) PNIL34, partial (57%) 426 e-120
TC209908 similar to UP|Q945B7 (Q945B7) Leucine zipper-containing... 130 8e-31
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 32 0.51
BF068779 31 0.87
TC230356 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (34%) 30 1.5
TC227920 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 30 1.9
TC233975 weakly similar to UP|Q9FGH8 (Q9FGH8) Similarity to sort... 29 3.3
TC211175 weakly similar to UP|Q9LIE8 (Q9LIE8) Similarity to cell... 29 3.3
TC228901 weakly similar to UP|Q9XHD3 (Q9XHD3) PHAP2B protein, pa... 29 4.3
TC231947 similar to UP|Q84NI9 (Q84NI9) Myb51 protein, partial (13%) 29 4.3
TC221428 similar to UP|BRH2_DROME (Q24256) Homeobox B-H2 protein... 29 4.3
TC227922 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like pro... 29 4.3
TC223581 28 5.7
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 28 5.7
TC207353 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 28 5.7
BG238010 similar to GP|15991300|dbj homeodomain transcription fa... 28 7.4
TC210712 28 7.4
TC225496 UP|Q6I687 (Q6I687) Protein disulfide isomerase-like pro... 28 7.4
TC214297 similar to UP|Q8S2S7 (Q8S2S7) Ethylene responsive eleme... 28 7.4
TC208197 similar to UP|Q9ZQT8 (Q9ZQT8) WREBP-2 protein, partial ... 28 7.4
>TC230339 homologue to UP|Q40093 (Q40093) PNIL34, partial (57%)
Length = 834
Score = 426 bits (1096), Expect = e-120
Identities = 203/212 (95%), Positives = 208/212 (97%)
Frame = +2
Query: 191 LTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDEN 250
LTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPE+QCYPIFKYFENWCQDEN
Sbjct: 2 LTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEFQCYPIFKYFENWCQDEN 181
Query: 251 RHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDM 310
RHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT FYEGIGLNTKEFDM
Sbjct: 182 RHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTAFYEGIGLNTKEFDM 361
Query: 311 HVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLI 370
HVIIETNRTTARIFPAVLDVENPEFKR+LDRMVEINEK++AVGESDDI VKNLKRIPL+
Sbjct: 362 HVIIETNRTTARIFPAVLDVENPEFKRKLDRMVEINEKLVAVGESDDIAVVKNLKRIPLV 541
Query: 371 AALASELLATYLMPPIESGSVDLAEFEPQLVY 402
AAL SELLATYLMPPIESGSVDLAEFEPQLVY
Sbjct: 542 AALVSELLATYLMPPIESGSVDLAEFEPQLVY 637
>TC209908 similar to UP|Q945B7 (Q945B7) Leucine zipper-containing protein,
partial (18%)
Length = 420
Score = 130 bits (328), Expect = 8e-31
Identities = 66/92 (71%), Positives = 76/92 (81%)
Frame = -2
Query: 43 TKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQT 102
T+P+ S++ + + RF +TDFDEM +L N INKNLN+ EFEALLQEFKTDYNQT
Sbjct: 275 TQPASSRATFSVRSS*VQ-RFSSTDFDEMASLLNF*INKNLNQDEFEALLQEFKTDYNQT 99
Query: 103 HFVRNKEFKEAADKLDGPLRQIFVEFLERSCT 134
HFVRNKEFKEAADKL+GPLRQIFVEFLERSCT
Sbjct: 98 HFVRNKEFKEAADKLEGPLRQIFVEFLERSCT 3
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 32.0 bits (71), Expect = 0.51
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Frame = +1
Query: 11 TNAAPKFSNPRT-ISHSKFSTVRMSATTTPPSSTKPSKKANKTS 53
T+ +P+ S+P + HS S S+T+ SSTKPS ++++ S
Sbjct: 52 TSPSPRISSPAPFLPHSPLSPASASSTSPTMSSTKPSPRSSRAS 183
>BF068779
Length = 408
Score = 31.2 bits (69), Expect = 0.87
Identities = 14/31 (45%), Positives = 20/31 (64%)
Frame = +1
Query: 14 APKFSNPRTISHSKFSTVRMSATTTPPSSTK 44
AP+ S P T + + FST R + T PPSS++
Sbjct: 232 APETSTPSTATTTTFSTKRTRSRTVPPSSSR 324
>TC230356 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (34%)
Length = 654
Score = 30.4 bits (67), Expect = 1.5
Identities = 19/55 (34%), Positives = 26/55 (46%)
Frame = +1
Query: 6 PISKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLT 60
P SK + P SNP S + +T S+++T PSS S +A S T T
Sbjct: 292 PASKPSKPEPHSSNPFAASQTPSTTSNTSSSSTTPSSATTSSRAPPGSSCATTTT 456
>TC227920 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (13%)
Length = 675
Score = 30.0 bits (66), Expect = 1.9
Identities = 17/59 (28%), Positives = 26/59 (43%), Gaps = 4/59 (6%)
Frame = -1
Query: 244 NWCQDENRHGDFFSAL----MKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFY 298
NWC+++ H F SA +P F N+ A L C + +DC+ D+Y
Sbjct: 261 NWCEEKQAHYYFISASKCSNRPPKPSFFNERLASL-----CNQQHSPACWHDCKNADYY 100
>TC233975 weakly similar to UP|Q9FGH8 (Q9FGH8) Similarity to sorting nexin,
partial (15%)
Length = 634
Score = 29.3 bits (64), Expect = 3.3
Identities = 19/54 (35%), Positives = 26/54 (47%)
Frame = +1
Query: 8 SKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTP 61
+K +N P P+T S S +R S T+ PPSS P + TS L +P
Sbjct: 370 TKPSNPPPPDPAPKTTSTSPSPILRRSRTSPPPSSPAP-PPSTPTSSPRALTSP 528
>TC211175 weakly similar to UP|Q9LIE8 (Q9LIE8) Similarity to cell wall-plasma
membrane linker protein, partial (3%)
Length = 609
Score = 29.3 bits (64), Expect = 3.3
Identities = 15/43 (34%), Positives = 22/43 (50%)
Frame = +1
Query: 5 KPISKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSK 47
KP S T+ AP +P S+ V S +T PP++ P+K
Sbjct: 190 KPPSPATHPAPPAKSPTLSPPSQSPAVSPSGSTPPPATNPPAK 318
>TC228901 weakly similar to UP|Q9XHD3 (Q9XHD3) PHAP2B protein, partial (8%)
Length = 699
Score = 28.9 bits (63), Expect = 4.3
Identities = 18/51 (35%), Positives = 22/51 (42%), Gaps = 9/51 (17%)
Frame = -2
Query: 200 FKPKFIFYATYLSEKIGYWRY---------ITIYRHLKANPEYQCYPIFKY 241
FKPKF+ IGYWR IT H KA C+ I+K+
Sbjct: 695 FKPKFVNSVXSAIINIGYWRLRTN*RKQRKITTMTH-KAGLNLACFQIYKF 546
>TC231947 similar to UP|Q84NI9 (Q84NI9) Myb51 protein, partial (13%)
Length = 756
Score = 28.9 bits (63), Expect = 4.3
Identities = 23/75 (30%), Positives = 34/75 (44%), Gaps = 2/75 (2%)
Frame = +1
Query: 11 TNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTS--IKETLLTPRFYTTDF 68
T+ P + + S S+ + TTT SST S +N S E LL P F +D
Sbjct: 322 TDEVPIIESNEILVPSAPSSTSPTTTTTTSSSTSTSASSNSNSSNFLEDLLLPDFEWSD- 498
Query: 69 DEMETLFNTEINKNL 83
D +T F+ N ++
Sbjct: 499 DYNDTNFDNNNNSSM 543
>TC221428 similar to UP|BRH2_DROME (Q24256) Homeobox B-H2 protein (Homeobox
BarH2 protein), partial (4%)
Length = 1188
Score = 28.9 bits (63), Expect = 4.3
Identities = 20/62 (32%), Positives = 27/62 (43%), Gaps = 4/62 (6%)
Frame = +2
Query: 11 TNAAPKFSNPRTISHSKFST----VRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTT 66
TN P S P+T HSK ST R A P S + + +A++ + L P T
Sbjct: 56 TNREPSLSEPKTTPHSKPSTPLTWTRWRAPKLPSSQSPLTPEASQLPQNKILEDPTH*TP 235
Query: 67 DF 68
F
Sbjct: 236 SF 241
>TC227922 similar to UP|Q7XJC6 (Q7XJC6) Callose synthase-like protein
(Fragment), partial (23%)
Length = 1119
Score = 28.9 bits (63), Expect = 4.3
Identities = 17/59 (28%), Positives = 25/59 (41%), Gaps = 4/59 (6%)
Frame = -3
Query: 244 NWCQDENRHGDFFSAL----MKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFY 298
NWC+++ H F SA +P F N+ A L C +DC+ D+Y
Sbjct: 532 NWCEEKQAHYYFISACKCSNRPPKPSFFNERLASL-----CNQQNSPARWHDCKNADYY 371
>TC223581
Length = 445
Score = 28.5 bits (62), Expect = 5.7
Identities = 17/54 (31%), Positives = 28/54 (51%)
Frame = +1
Query: 15 PKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDF 68
P +NP+ + +T+R S +PP S PS KA+ T+++ + P T F
Sbjct: 64 PTKTNPKPSPAAITTTLRRSHNPSPPPSMAPS-KASPTTLRRSRTPPSASLTRF 222
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 28.5 bits (62), Expect = 5.7
Identities = 19/56 (33%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Frame = +2
Query: 6 PISKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKK-ANKTSIKETLLT 60
P+S A S P+T+S S +A T+PP+ST P ++ A K+ T T
Sbjct: 671 PLSGLRPAPSTASTPKTLSPSS------TAPTSPPASTPPPRRPARKSPTPTTTFT 820
>TC207353 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (58%)
Length = 898
Score = 28.5 bits (62), Expect = 5.7
Identities = 22/68 (32%), Positives = 29/68 (42%), Gaps = 7/68 (10%)
Frame = +1
Query: 6 PISKFTNAAPKFSNPRTISHSKFSTV------RMSATTTPPSSTKPSKKANKTSIKETLL 59
P S T P S P T++ ++F T R S+ PP S P+ +S LL
Sbjct: 133 PPSTTTATEPSSSPPATMAPARFGTPEPAIS*RPSSKIRPPLSPSPNSPPTASSFSPPLL 312
Query: 60 -TPRFYTT 66
TP Y T
Sbjct: 313 TTPLSYGT 336
>BG238010 similar to GP|15991300|dbj homeodomain transcription factor KNAT6
{Arabidopsis thaliana}, partial (19%)
Length = 478
Score = 28.1 bits (61), Expect = 7.4
Identities = 24/74 (32%), Positives = 34/74 (45%)
Frame = +1
Query: 6 PISKFTNAAPKFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYT 65
P T P ++P SH KFS ATTTP S+KP K+ + T L T
Sbjct: 232 PSPPATKPNPTLTSP---SHQKFS-----ATTTPLLSSKP-----KSPLTLTTLVSSKPT 372
Query: 66 TDFDEMETLFNTEI 79
++ +E L N+ +
Sbjct: 373 SNVRRLERLQNSRV 414
>TC210712
Length = 776
Score = 28.1 bits (61), Expect = 7.4
Identities = 21/84 (25%), Positives = 35/84 (41%)
Frame = -3
Query: 259 LMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNR 318
L+ P N +K+ L S F +S + DCQ+T + + N I R
Sbjct: 564 LVTLMPSDTNSFKSPLISSLFNMSRNL-FDTPDCQKTTMAQLLSTNLTSNSREKISWEER 388
Query: 319 TTARIFPAVLDVENPEFKRRLDRM 342
+ PAV+ + NP K + ++
Sbjct: 387 SKLFSMPAVIPILNPHKKTKCSQL 316
>TC225496 UP|Q6I687 (Q6I687) Protein disulfide isomerase-like protein,
complete
Length = 2048
Score = 28.1 bits (61), Expect = 7.4
Identities = 12/43 (27%), Positives = 22/43 (50%)
Frame = +1
Query: 88 FEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLE 130
F AL ++ ++DY+ H + K + GP+ ++F F E
Sbjct: 634 FSALAEKLRSDYDFGHTLNAKHLPRGESSVSGPVVRLFKPFDE 762
>TC214297 similar to UP|Q8S2S7 (Q8S2S7) Ethylene responsive element binding
factor 4-like protein, partial (51%)
Length = 996
Score = 28.1 bits (61), Expect = 7.4
Identities = 21/50 (42%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Frame = +1
Query: 6 PISKFTNAAPKFSNPRTISHSKFSTVRMSATTT-PPSSTKPSKKANKTSI 54
P ++FT + S P SHS + VR AT T PPSS+ S K + SI
Sbjct: 499 PCARFTFSTGPTS*PSP-SHSDSNRVRSRATPTRPPSSSIASPKESSISI 645
>TC208197 similar to UP|Q9ZQT8 (Q9ZQT8) WREBP-2 protein, partial (57%)
Length = 1002
Score = 28.1 bits (61), Expect = 7.4
Identities = 23/101 (22%), Positives = 41/101 (39%), Gaps = 1/101 (0%)
Frame = +1
Query: 16 KFSNPRTISHSKFSTVRMSATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLF 75
K N H K ++V ++ TTPPS + + + R Y +D E +
Sbjct: 169 KLKNHIASHHEKNASVDVTKYTTPPSEKQTKTSKPSGGAYGSASSDRPYACPYDGCEKAY 348
Query: 76 NTEINKNLN-EHEFEALLQEFKTDYNQTHFVRNKEFKEAAD 115
E L+ + E L+ + ++ Q + + E EA+D
Sbjct: 349 IHEYKLRLHLKREHPGLMADENAEHAQANV--DNEMDEASD 465
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,640,574
Number of Sequences: 63676
Number of extensions: 233484
Number of successful extensions: 1569
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1550
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1566
length of query: 402
length of database: 12,639,632
effective HSP length: 99
effective length of query: 303
effective length of database: 6,335,708
effective search space: 1919719524
effective search space used: 1919719524
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0098e.6


