

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
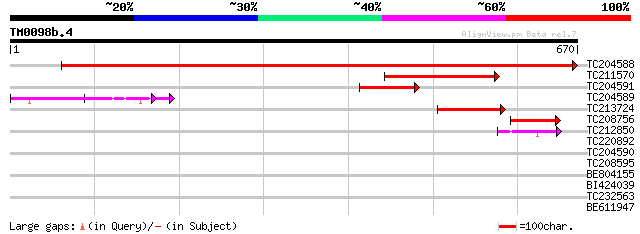
Query= TM0098b.4
(670 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 1089 0.0
TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 194 1e-49
TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydr... 124 1e-28
TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosida... 122 4e-28
TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-... 87 3e-17
TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 59 5e-09
TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofur... 56 4e-08
TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-rel... 41 0.001
TC204590 36 0.048
TC208595 weakly similar to UP|Q9LIQ0 (Q9LIQ0) Arabidopsis thalia... 33 0.53
BE804155 32 0.91
BI424039 homologue to GP|23093676|gb| CG32071-PA {Drosophila mel... 32 0.91
TC232563 weakly similar to GB|AAQ89617.1|37202004|BT010595 At3g1... 32 0.91
BE611947 homologue to GP|23476279|gb| myb-like transcription fac... 29 7.7
>TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(84%)
Length = 2003
Score = 1089 bits (2816), Expect = 0.0
Identities = 526/611 (86%), Positives = 567/611 (92%), Gaps = 2/611 (0%)
Frame = +2
Query: 62 GAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVL 121
G GLWAELVSNRGFEAGGPN PSNIDPWSIIGNES IN+ETDRTSCFDRNK+ALRLEVL
Sbjct: 8 GTRGLWAELVSNRGFEAGGPNTPSNIDPWSIIGNESLINIETDRTSCFDRNKVALRLEVL 187
Query: 122 CDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGV--LA 179
CDSKGDNICPADGVGV+NPGFWGMNIEQGKKYKVVFYVRST SL+LTVSLTGSNGV LA
Sbjct: 188 CDSKGDNICPADGVGVYNPGFWGMNIEQGKKYKVVFYVRSTESLNLTVSLTGSNGVGNLA 367
Query: 180 SNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGF 239
S+VITGSA DFSNW KVE +L AKAT+HNSRLQLTTT KGVIWLDQVSAMPLDTYKG GF
Sbjct: 368 SSVITGSALDFSNWTKVEKILVAKATDHNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGF 547
Query: 240 RSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDG 299
R++LV ML LKPRFIRFPGGCFVEGEWLRNAFRWK SIGPWEERPGHFGDVW YWTDDG
Sbjct: 548 RTDLVEMLTALKPRFIRFPGGCFVEGEWLRNAFRWKTSIGPWEERPGHFGDVWMYWTDDG 727
Query: 300 LGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGS 359
LGY+EFLQLAEDLG PIWVFNNGVSHND+VDTSAVLP VQEALDG+EFARGD TSKWGS
Sbjct: 728 LGYYEFLQLAEDLGTAPIWVFNNGVSHNDQVDTSAVLPLVQEALDGLEFARGDRTSKWGS 907
Query: 360 IRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLD 419
+RA+MGHP PF+L+YVAVGNEDCGKKNYRGNYLKFYSAI+SAYPDIQIISNCDGSSRPLD
Sbjct: 908 LRAAMGHPAPFDLRYVAVGNEDCGKKNYRGNYLKFYSAIKSAYPDIQIISNCDGSSRPLD 1087
Query: 420 HPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGF 479
HPAD+YDYH+YTNANDMFSR++ F+ R+GPKAFVSEYAVTG DAGTGSLLAA+AEAGF
Sbjct: 1088HPADMYDYHVYTNANDMFSRANAFDHTSRNGPKAFVSEYAVTGKDAGTGSLLAALAEAGF 1267
Query: 480 LLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATL 539
L+GLEKNSD+++M SYAPLFVNANDRRWNPDAIVFNSFQ YGTPSYWVQLFFSESSGATL
Sbjct: 1268LIGLEKNSDVIQMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWVQLFFSESSGATL 1447
Query: 540 LNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSG 599
L++SLQ SS+SL+ASAI +Q+S DKKNYIRIKVVNFG VNLKISL+GL+PNSLQ SG
Sbjct: 1448LSTSLQAPSSNSLVASAITFQNSVDKKNYIRIKVVNFGTTAVNLKISLDGLEPNSLQLSG 1627
Query: 600 STRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSNLKM 659
ST+TVLTSANVMDENSFSQPKKVVPIQSLLQNVGK+MNV VPP SF+SFDL+K+SS LKM
Sbjct: 1628STKTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKDMNVTVPPRSFTSFDLVKQSSTLKM 1807
Query: 660 VGGGSSTRSSI 670
G SSTRSSI
Sbjct: 1808AGSDSSTRSSI 1840
>TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(20%)
Length = 414
Score = 194 bits (492), Expect = 1e-49
Identities = 96/136 (70%), Positives = 113/136 (82%)
Frame = +3
Query: 443 FNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNA 502
F+ RSGPKAFVSEYAV +DA G+LLAA+AEA FL+GLEKNSDI +MVSYAPLF+N
Sbjct: 6 FDNTSRSGPKAFVSEYAVK-SDAANGNLLAAVAEAAFLIGLEKNSDIGEMVSYAPLFLNI 182
Query: 503 NDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSS 562
ND+RW PDAIVFNS+Q YGTPSYWVQ F ESSGAT ++S+L T+SS+ L AS I WQ S
Sbjct: 183 NDKRWIPDAIVFNSYQVYGTPSYWVQKLFIESSGATFIDSTLSTTSSNKLAASVIIWQDS 362
Query: 563 ADKKNYIRIKVVNFGA 578
+DK+NY+RIK VNFGA
Sbjct: 363 SDKRNYLRIKAVNFGA 410
>TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydrogenase ,
partial (84%)
Length = 1604
Score = 124 bits (311), Expect = 1e-28
Identities = 57/71 (80%), Positives = 66/71 (92%)
Frame = +2
Query: 414 SSRPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAA 473
+SRPLDHPAD+YDYH+YTNANDMFSR++ F+ R+GPKAFVSEYAVTG DAGTGSLLAA
Sbjct: 1391 ASRPLDHPADMYDYHVYTNANDMFSRANAFDHTSRNGPKAFVSEYAVTGKDAGTGSLLAA 1570
Query: 474 IAEAGFLLGLE 484
+AEAGFL+GLE
Sbjct: 1571 LAEAGFLIGLE 1603
>TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosidase, partial
(14%)
Length = 808
Score = 122 bits (307), Expect = 4e-28
Identities = 85/177 (48%), Positives = 102/177 (57%), Gaps = 4/177 (2%)
Frame = +3
Query: 1 MGLYKASFIVAALQLCIAVSLC----AIKVNADLNATLTVDAHETSGKPISETLFGLFFE 56
MG KA V+ L L I V L A K +ADL TLTV+A + SGKPI ETLFG+FFE
Sbjct: 222 MGSSKALCSVSLLHLFIVVCLVFQCFATKKHADLTTTLTVNASDASGKPIPETLFGIFFE 401
Query: 57 EINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIAL 116
EINHAGAGGLWAELVSNRGFEAGGPN PSNI ++ ++ R ++
Sbjct: 402 EINHAGAGGLWAELVSNRGFEAGGPNTPSNIALGQLLEMNH**ILKLIAPLALTRTRVHS 581
Query: 117 RLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTG 173
+ + +I PADGV V GMNIEQ ++ FY R L + SLTG
Sbjct: 582 DWRCM-*HQX*HIWPADGV-VAITLVLGMNIEQXEEXXXGFYCRQLDHL-IYXSLTG 743
Score = 63.9 bits (154), Expect = 2e-10
Identities = 47/112 (41%), Positives = 57/112 (49%), Gaps = 6/112 (5%)
Frame = +2
Query: 89 PWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIE 148
PWSIIGNES IN+ETDRTSCFD K ALRLEV ++ G G +NPG
Sbjct: 497 PWSIIGNESLINIETDRTSCFDPXKGALRLEVYVTPXVTHLAR*WG-GGYNPG------- 652
Query: 149 QGKKY------KVVFYVRSTGSLDLTVSLTGSNGVLASNVITGSASDFSNWK 194
G +Y + STGSL+L + SNGV + F NW+
Sbjct: 653 SGHEY*AXGGNXXXLLLSSTGSLNLXF-INWSNGVGNWFIWDTGLHLFXNWQ 805
>TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-1, partial
(10%)
Length = 595
Score = 86.7 bits (213), Expect = 3e-17
Identities = 46/81 (56%), Positives = 55/81 (67%)
Frame = +3
Query: 506 RWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADK 565
RW+PDAIVFNS Q Y TPSY VQ F ES+GAT L S LQT S+ ASAI W++ DK
Sbjct: 141 RWSPDAIVFNSNQVYETPSYRVQFMFRESNGATFLKSQLQTPDPDSVDASAILWKNLQDK 320
Query: 566 KNYIRIKVVNFGANTVNLKIS 586
K Y++IKV+ + LKIS
Sbjct: 321 KTYLKIKVIQ*RWHQSVLKIS 383
>TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(5%)
Length = 544
Score = 59.3 bits (142), Expect = 5e-09
Identities = 32/59 (54%), Positives = 41/59 (69%)
Frame = +1
Query: 593 NSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLL 651
+++Q GST TVLTS NVMDENSF +PKKVVP S L+ K + V++P +S S D L
Sbjct: 4 SNVQQYGSTITVLTSTNVMDENSFLEPKKVVPQTSPLEIDDKHLIVLLPSYSVISLDFL 180
>TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofuranosidase,
partial (3%)
Length = 558
Score = 56.2 bits (134), Expect = 4e-08
Identities = 38/111 (34%), Positives = 52/111 (46%), Gaps = 35/111 (31%)
Frame = +2
Query: 577 GANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPKKV-------------- 622
G+N +N KISLNG + S + +T+TVLTS N +DENS + P V
Sbjct: 11 GSNQINFKISLNGFE--SSNATKATKTVLTSKNALDENSLANPTNVHITSFILFLLL*TN 184
Query: 623 ---------------------VPIQSLLQNVGKEMNVVVPPHSFSSFDLLK 652
VP +S LQ+ E+N ++PP S + FDLLK
Sbjct: 185 NTYNN*LS*HLMFICLFCTQIVPKRSPLQSANNEINDILPPVSLTVFDLLK 337
>TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-related
protein, partial (5%)
Length = 647
Score = 41.2 bits (95), Expect = 0.001
Identities = 21/38 (55%), Positives = 28/38 (73%), Gaps = 2/38 (5%)
Frame = +3
Query: 146 NIEQGKKYKVVFYVRSTGSLDLTVSLTGS--NGVLASN 181
NIE+G+KYKVVFY ++ G+++L VS GS LASN
Sbjct: 129 NIEKGQKYKVVFYAKADGAINLDVSFVGSEKGEKLASN 242
>TC204590
Length = 290
Score = 36.2 bits (82), Expect = 0.048
Identities = 18/23 (78%), Positives = 19/23 (82%)
Frame = +2
Query: 648 FDLLKESSNLKMVGGGSSTRSSI 670
FDLLK+SSNLKM G SST SSI
Sbjct: 2 FDLLKQSSNLKMAGSDSSTWSSI 70
>TC208595 weakly similar to UP|Q9LIQ0 (Q9LIQ0) Arabidopsis thaliana genomic
DNA, chromosome 3, BAC clone:F14O13, partial (20%)
Length = 979
Score = 32.7 bits (73), Expect = 0.53
Identities = 25/72 (34%), Positives = 37/72 (50%)
Frame = -3
Query: 529 LFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLN 588
LF SSG SS + SSSL+ S +++ SSA ++ + + F N +L
Sbjct: 227 LF*LPSSGIFSSFSSSPSGRSSSLVFSNLSFSSSASGRSSSLVFSI-FSFNAFSLAAKNP 51
Query: 589 GLDPNSLQPSGS 600
LDP++ QP GS
Sbjct: 50 PLDPHASQPLGS 15
>BE804155
Length = 409
Score = 32.0 bits (71), Expect = 0.91
Identities = 24/66 (36%), Positives = 29/66 (43%)
Frame = +3
Query: 593 NSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLK 652
NSL PS T T ++ + SF++P K P LQ N PP FSS L
Sbjct: 195 NSLYPSSPHITTSTVSSTRSKPSFTRPSKPPP----LQLPPSRTNSWSPPPQFSSAHLKT 362
Query: 653 ESSNLK 658
S LK
Sbjct: 363 SLSRLK 380
>BI424039 homologue to GP|23093676|gb| CG32071-PA {Drosophila melanogaster},
partial (6%)
Length = 380
Score = 32.0 bits (71), Expect = 0.91
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Frame = -1
Query: 583 LKISLNGLDPNS-LQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSL 628
LK L GL PN+ P+ STR ++ + MDE+ +P PI +
Sbjct: 290 LKSQLEGLGPNTETSPATSTRGTMSKKSFMDEDDSGRPHLTPPINGV 150
>TC232563 weakly similar to GB|AAQ89617.1|37202004|BT010595 At3g10650
{Arabidopsis thaliana;} , partial (4%)
Length = 602
Score = 32.0 bits (71), Expect = 0.91
Identities = 25/103 (24%), Positives = 47/103 (45%), Gaps = 3/103 (2%)
Frame = +3
Query: 516 SFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNY---IRIK 572
S ++G+ S +FF SS T +NSS SS+ + ++ ++ + + +
Sbjct: 150 SSTAFGSSSAGSGIFFFSSSAMTTVNSSQSQSSAVGASSGSVLGAQASFTSGFATSTQTQ 329
Query: 573 VVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENS 615
V+FG++ + L+G P S S + + SA + NS
Sbjct: 330 SVSFGSSASSSSFGLSGNTPFSSSSSFPSSSPAASAAFLSGNS 458
>BE611947 homologue to GP|23476279|gb| myb-like transcription factor 1
{Gossypium hirsutum}, partial (31%)
Length = 400
Score = 28.9 bits (63), Expect = 7.7
Identities = 20/92 (21%), Positives = 40/92 (42%)
Frame = +1
Query: 531 FSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGL 590
F+E ++N + +LIA+ + ++ + KNY + K+ G+
Sbjct: 100 FTEEEDELIINLHSLLGNKWALIAARLPGRTDNEIKNYWNTHIKR--------KLYSRGI 255
Query: 591 DPNSLQPSGSTRTVLTSANVMDENSFSQPKKV 622
DP + +P ++ T T+A S + KK+
Sbjct: 256 DPQTHRPLNASATPATTATATAVPSANSSKKI 351
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,457,844
Number of Sequences: 63676
Number of extensions: 440552
Number of successful extensions: 2110
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 2085
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2104
length of query: 670
length of database: 12,639,632
effective HSP length: 104
effective length of query: 566
effective length of database: 6,017,328
effective search space: 3405807648
effective search space used: 3405807648
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0098b.4


