

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
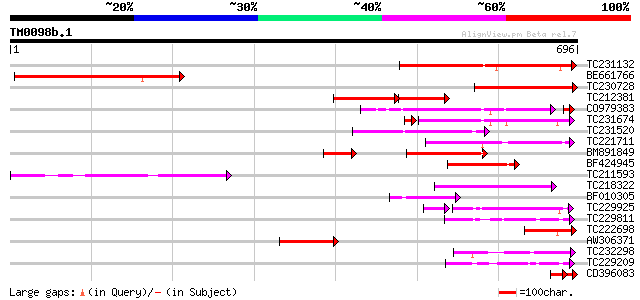
Query= TM0098b.1
(696 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231132 weakly similar to UP|Q93YF5 (Q93YF5) SET-domain-contain... 268 5e-72
BE661766 similar to GP|15485584|emb SET-domain-containing protei... 253 2e-67
TC230728 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing pro... 241 1e-63
TC212381 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing pro... 144 2e-60
CO979383 189 4e-49
TC231674 similar to UP|SUV6_ARATH (Q8VZ17) Histone-lysine N-meth... 164 8e-42
TC231520 weakly similar to UP|Q8L821 (Q8L821) SET domain-contain... 134 1e-31
TC221711 weakly similar to UP|Q84XG3 (Q84XG3) SET domain protein... 131 1e-30
BM891849 130 2e-30
BF424945 similar to GP|13517747|gb| SUVH3 {Arabidopsis thaliana}... 124 2e-28
TC211593 similar to UP|SUV1_ARATH (Q9FF80) Histone-lysine N-meth... 118 8e-27
TC218322 similar to UP|SUV9_ARATH (Q9T0G7) Probable histone-lysi... 109 5e-24
BF010305 weakly similar to GP|13517745|gb| SUVH2 {Arabidopsis th... 92 6e-19
TC229925 similar to UP|Q946J2 (Q946J2) Suppressor of variegation... 73 9e-16
TC229811 similar to UP|Q9FIH7 (Q9FIH7) Similarity to SET-domain ... 78 1e-14
TC222698 similar to UP|SUV8_ARATH (Q9C5P0) Histone-lysine N-meth... 75 1e-13
AW306371 similar to GP|13517751|gb| SUVH5 {Arabidopsis thaliana}... 64 3e-10
TC232298 similar to GB|AAO64916.1|29029074|BT005981 At5g53430 {A... 62 9e-10
TC229209 similar to UP|Q9C5X4 (Q9C5X4) Trithorax-like protein 1,... 61 1e-09
CD396083 43 2e-08
>TC231132 weakly similar to UP|Q93YF5 (Q93YF5) SET-domain-containing protein,
partial (24%)
Length = 1134
Score = 268 bits (686), Expect = 5e-72
Identities = 132/233 (56%), Positives = 162/233 (68%), Gaps = 16/233 (6%)
Frame = +2
Query: 479 CQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLE 538
C PG C C ++N G PY+ G+L K +V+ECGP+C C P+C+NRVSQ GLK +E
Sbjct: 5 CVPGDLNCSCIRRNEGDFPYTGNGILVSRKPLVHECGPTCQCFPNCKNRVSQTGLKHPME 184
Query: 539 VFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL-- 596
VFRTK +GWGLRS DPIRAGTFICEYAGEV+D +V +L E D+Y+FD+TRIY Q
Sbjct: 185 VFRTKDRGWGLRSLDPIRAGTFICEYAGEVVDRGKVSQLVKEG-DEYVFDTTRIYDQFKW 361
Query: 597 --------EVFSSD-VEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEA 647
E+ S+D E +P PL ITA+N GNVARFMNHSC+PNV W+PVV E N++
Sbjct: 362 NYEPRLLEEIGSNDSTEDYAMPYPLIITAKNIGNVARFMNHSCSPNVFWQPVVYEENNQS 541
Query: 648 DLHVAFYAIRHIPPMMELTYDYGIVLP-----LKVGQKKKKCLCGSVKCRGYF 695
LHVAF+A+RHIPPM ELTYDYG+ + +KKCLCGS KCRG F
Sbjct: 542 YLHVAFFALRHIPPMTELTYDYGLAQSDHAEGSSAAKGRKKCLCGSSKCRGSF 700
>BE661766 similar to GP|15485584|emb SET-domain-containing protein {Nicotiana
tabacum}, partial (4%)
Length = 875
Score = 253 bits (646), Expect = 2e-67
Identities = 136/220 (61%), Positives = 166/220 (74%), Gaps = 11/220 (5%)
Frame = +3
Query: 6 GQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGV 65
GQ PA+ SFDK+RVLNVKPLRTLVPVFPSPSNP+SS+TPQGGAPFVCVSPSGPFPSGV
Sbjct: 6 GQGTTPASESFDKTRVLNVKPLRTLVPVFPSPSNPASSSTPQGGAPFVCVSPSGPFPSGV 185
Query: 66 APFYPFFVSPESQRLSEQNAQTPTAQR--AAPISAAVPINSFRTPTGATNGDVGSSRR-- 121
APFYPFF+S ESQRLSEQNAQTP+ QR AAP+S AVPINSFRTPTGA NGDVGSS++
Sbjct: 186 APFYPFFISSESQRLSEQNAQTPSGQRVPAAPVSTAVPINSFRTPTGAANGDVGSSQKNT 365
Query: 122 KSRGQLPEDDNF--VDLSEVDGEGGTGDGK-RRKPQKRIREKR----CSSDVDPDAVANE 174
+SRG + E+D + V++ E+D + GTG G+ +RK K+ + ++ S DVDPDAVA +
Sbjct: 366 RSRGWVTEEDGYSNVEIEEIDADEGTGTGRSKRKSNKKTKARQSGGSVSGDVDPDAVAAD 545
Query: 175 ILKTINPGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQI 214
ILK++N VF++L + AY + R GQI
Sbjct: 546 ILKSLNALVFDVLKSTRR*XGSSAYHS*YMK*CXRSFGQI 665
Score = 34.3 bits (77), Expect = 0.19
Identities = 28/85 (32%), Positives = 44/85 (50%), Gaps = 4/85 (4%)
Frame = +2
Query: 200 TLMIYEVMRRKLGQIDEKAKGSHS--GAKRPDLKAGTLM-NTKGIRANSRKR-IGVVPGV 255
+LMIYEVM +KL + + + + DLK + KGIR + + + + G+
Sbjct: 620 SLMIYEVMPKKLWTNWKNSNRAXNFWNQNGLDLKGRCQL*MNKGIRTHRXETXLELXLGL 799
Query: 256 EVGDIFFFRFELCLVGLHAPSMAGI 280
+ G+I F +FE GLHAP G+
Sbjct: 800 KXGEISFSKFEWA*XGLHAPLWLGM 874
>TC230728 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing protein,
partial (11%)
Length = 774
Score = 241 bits (614), Expect = 1e-63
Identities = 109/126 (86%), Positives = 119/126 (93%)
Frame = +2
Query: 571 NARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSC 630
+AR+EEL G+NEDDYI DSTRIYQQLEVF D EAPKIPSPLYI+A+NEGNVARFMNH C
Sbjct: 8 SARMEELGGDNEDDYICDSTRIYQQLEVFPGDTEAPKIPSPLYISAKNEGNVARFMNHCC 187
Query: 631 TPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVK 690
+PNVLWRPV+RENKNE+DLH+AFYAIRHIPPMMELTYDYG VLPLKVGQ+KKKCLCGSVK
Sbjct: 188 SPNVLWRPVIRENKNESDLHIAFYAIRHIPPMMELTYDYGAVLPLKVGQRKKKCLCGSVK 367
Query: 691 CRGYFC 696
CRGYFC
Sbjct: 368 CRGYFC 385
>TC212381 similar to UP|Q93YF5 (Q93YF5) SET-domain-containing protein,
partial (16%)
Length = 435
Score = 144 bits (362), Expect(2) = 2e-60
Identities = 68/81 (83%), Positives = 70/81 (85%)
Frame = +3
Query: 398 PGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPT 457
P QPQAYMIWKSI QWT+KSASR GVILPDLTSGAE +PVCLVNDVDNEKGPAYFTY PT
Sbjct: 6 PEQPQAYMIWKSIQQWTEKSASRAGVILPDLTSGAENVPVCLVNDVDNEKGPAYFTYIPT 185
Query: 458 LKNLNRLAPVESSEGCTCNGG 478
LKNL APVESS GC C GG
Sbjct: 186 LKNLRPTAPVESSTGCPCVGG 248
Score = 107 bits (268), Expect(2) = 2e-60
Identities = 48/63 (76%), Positives = 52/63 (82%)
Frame = +1
Query: 478 GCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRL 537
GCQ + C C QKNGGYLPYS+A LLADLKSV+YECGPSC CP +CRNRVSQ GLK RL
Sbjct: 247 GCQSKNFNCPCIQKNGGYLPYSSALLLADLKSVIYECGPSCQCPSNCRNRVSQSGLKFRL 426
Query: 538 EVF 540
EVF
Sbjct: 427 EVF 435
>CO979383
Length = 834
Score = 189 bits (480), Expect(2) = 4e-49
Identities = 98/253 (38%), Positives = 147/253 (57%), Gaps = 14/253 (5%)
Frame = -3
Query: 431 GAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQ 490
G E++P+C VN +D+E P F Y ++ N V +EGC C GC KC C
Sbjct: 832 GKERIPICAVNTIDDENPPP-FNYITSMIYPN--CHVLPAEGCDCTNGCSD-LEKCSCVV 665
Query: 491 KNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLR 550
KNGG +P++ + K +VYECGP+C CP +C NRVSQ G+K +LE+F+T +GWG+R
Sbjct: 664 KNGGEIPFNHNEAIVQAKPLVYECGPTCKCPSTCHNRVSQLGIKFQLEIFKTDTRGWGVR 485
Query: 551 SWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFD--------------STRIYQQL 596
S + I +G+FICEY GE++++ E+ +G D+Y+FD ST
Sbjct: 484 SLNSIPSGSFICEYIGELLEDKEAEQRTG--NDEYLFDIGNNYSNSTLWDDLSTLTTLMP 311
Query: 597 EVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAI 656
+ ++ E K I A GN+ RF+NHSC+PN++ + V+ ++ + H+ F+A
Sbjct: 310 DAHTASCEVVK-DGGFTIDAAQFGNLGRFINHSCSPNLIAQNVLYDHHDTRMPHIMFFAA 134
Query: 657 RHIPPMMELTYDY 669
+IPP+ ELTYDY
Sbjct: 133 DNIPPLQELTYDY 95
Score = 24.6 bits (52), Expect(2) = 4e-49
Identities = 9/14 (64%), Positives = 9/14 (64%)
Frame = -1
Query: 680 KKKKCLCGSVKCRG 693
K K C CGSV C G
Sbjct: 51 KXKXCYCGSVDCTG 10
>TC231674 similar to UP|SUV6_ARATH (Q8VZ17) Histone-lysine
N-methyltransferase, H3 lysine-9 specific 6 (Histone
H3-K9 methyltransferase 6) (H3-K9-HMTase 6) (Suppressor
of variegation 3-9 homolog 6) (Su(var)3-9 homolog 6) ,
partial (24%)
Length = 755
Score = 164 bits (414), Expect(2) = 8e-42
Identities = 83/206 (40%), Positives = 122/206 (58%), Gaps = 14/206 (6%)
Frame = +1
Query: 502 GLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFI 561
G + K +VYECGP+C CP +C NRVSQ G+K +LE+F+T +GWG+RS + I +G+FI
Sbjct: 61 GAIVQAKPLVYECGPTCKCPSTCHNRVSQLGIKFQLEIFKTDTRGWGVRSLNSIPSGSFI 240
Query: 562 CEYAGEVIDNARVEELSGENEDDYIFD------STRIYQQLEVFSSDVEAPKI----PSP 611
CEY GE++++ E+ +G D+Y+FD ++ ++ L DV
Sbjct: 241 CEYIGELLEDKEAEQRTG--NDEYLFDIGNNYSNSALWDDLSTLMPDVHTTSCEVVKDGG 414
Query: 612 LYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGI 671
I A GNV RF+NHSC+PN++ + V+ +N + H+ F+A +IPP+ ELTYDY
Sbjct: 415 FTIDAAQFGNVGRFINHSCSPNLIAQNVLYDNHDTRMPHIMFFAADNIPPLQELTYDYNY 594
Query: 672 ----VLPLKVGQKKKKCLCGSVKCRG 693
+ KKK C CGSV+C G
Sbjct: 595 EIDQIRDSGGNIKKKYCHCGSVECTG 672
Score = 25.8 bits (55), Expect(2) = 8e-42
Identities = 8/15 (53%), Positives = 11/15 (73%)
Frame = +3
Query: 485 KCGCTQKNGGYLPYS 499
KC C KNGG +P++
Sbjct: 9 KCSCVVKNGGEIPFN 53
>TC231520 weakly similar to UP|Q8L821 (Q8L821) SET domain-containing protein
SET118, partial (24%)
Length = 565
Score = 134 bits (337), Expect = 1e-31
Identities = 71/170 (41%), Positives = 96/170 (55%), Gaps = 3/170 (1%)
Frame = +1
Query: 422 GVILPDLTSGAEKLPVCLVNDVDNEK-GPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQ 480
G++ D+T G E +P+ N VD+ P FTY +K + ++ GC C G C
Sbjct: 58 GLVCEDITGGQEDMPIPATNLVDDPPVPPTGFTYCKFVKVAKNVKLPMNATGCECKGICN 237
Query: 481 PGSHKCGCTQKNGGYLPYSA--AGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLE 538
+ C C +NG PY + G L + K VV+ECGP C C P C NR SQ GL+ RLE
Sbjct: 238 DPT-TCACALRNGSDFPYVSRDGGRLVEAKDVVFECGPECGCGPGCVNRTSQRGLRYRLE 414
Query: 539 VFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFD 588
VFRT KGW +RSWD I +G +CEY G + AR E++ E++YIF+
Sbjct: 415 VFRTAKKGWAVRSWDFIPSGAPVCEYTGIL---ARAEDMDSVLENNYIFE 555
>TC221711 weakly similar to UP|Q84XG3 (Q84XG3) SET domain protein SDG117,
partial (12%)
Length = 1208
Score = 131 bits (329), Expect = 1e-30
Identities = 74/187 (39%), Positives = 101/187 (53%), Gaps = 4/187 (2%)
Frame = +1
Query: 511 VYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVID 570
VYEC C C SC NRV Q G++++LEVF+T+ KGW +R+ + I GTF+CEY GEV+D
Sbjct: 1 VYECNHMCRCNKSCPNRVLQNGVRVKLEVFKTEKKGWAVRAGEAILRGTFVCEYIGEVLD 180
Query: 571 NARVEELS---GENEDDYIFD-STRIYQQLEVFSSDVEAPKIPSPLYITARNEGNVARFM 626
+ G Y++D R+ + + I A GNV+RF+
Sbjct: 181 VQEARDRRKRYGAEHCSYLYDIDARVNDMGRLIEEQAQ-------YVIDATKFGNVSRFI 339
Query: 627 NHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLC 686
NHSC+PN++ V+ E+ + H+ FYA R I ELTYDY L G CLC
Sbjct: 340 NHSCSPNLVNHQVLVESMDCERAHIGFYASRDIALGEELTYDYQYELMPGEG---SPCLC 510
Query: 687 GSVKCRG 693
S+KCRG
Sbjct: 511 ESLKCRG 531
>BM891849
Length = 421
Score = 130 bits (328), Expect = 2e-30
Identities = 61/99 (61%), Positives = 77/99 (77%)
Frame = +3
Query: 488 CTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGW 547
C Q+N G PY+A G+L K +V+ECGP C C P+C+NRVSQ GLK ++EVF+TK +GW
Sbjct: 126 CIQRNEGDFPYTANGVLVSRKPLVHECGPLCKCFPNCKNRVSQTGLKHQMEVFKTKDRGW 305
Query: 548 GLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYI 586
GLRS DPIRAGTFICEYAGEVID A+V + G +D+Y+
Sbjct: 306 GLRSLDPIRAGTFICEYAGEVIDIAKVNKNRG-YDDEYV 419
Score = 50.1 bits (118), Expect = 3e-06
Identities = 22/42 (52%), Positives = 30/42 (71%), Gaps = 2/42 (4%)
Frame = +1
Query: 386 GFNVFKYKLVRLPGQPQAYMIWKSILQW--TDKSASRVGVIL 425
G VFKYK VRLPGQ A+ +WKS+ +W + ++SR G+IL
Sbjct: 1 GGGVFKYKFVRLPGQSSAFAVWKSVQKWKMSSSTSSRTGIIL 126
>BF424945 similar to GP|13517747|gb| SUVH3 {Arabidopsis thaliana}, partial
(4%)
Length = 266
Score = 124 bits (310), Expect = 2e-28
Identities = 63/89 (70%), Positives = 71/89 (78%)
Frame = +2
Query: 538 EVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLE 597
EVFRTK KGWGLRSWD IRAGTFICEYAGEVI +ARVEEL G+N++D IFDST IY+QLE
Sbjct: 2 EVFRTKNKGWGLRSWDSIRAGTFICEYAGEVIYSARVEELGGDNDNDCIFDSTHIYRQLE 181
Query: 598 VFSSDVEAPKIPSPLYITARNEGNVARFM 626
VFS D + SP +I R GNV+RFM
Sbjct: 182 VFSYDTDRHICHSP-FIYQRK*GNVSRFM 265
>TC211593 similar to UP|SUV1_ARATH (Q9FF80) Histone-lysine
N-methyltransferase, H3 lysine-9 specific 1 (Histone
H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor
of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) ,
partial (5%)
Length = 791
Score = 118 bits (296), Expect = 8e-27
Identities = 85/273 (31%), Positives = 118/273 (43%), Gaps = 1/273 (0%)
Frame = +2
Query: 1 MDHNLGQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGP 60
M+ Q+ P +G DK+R++ KPLR+L PV P S S G
Sbjct: 95 MEEGSCQNSVPGSGFVDKTRIVEAKPLRSLAPVLPKSLQSSLS---------------GR 229
Query: 61 FPSGVAPFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVPINSFRTPTGATNGDVGSSR 120
+PS PF F ESQ +P PI S+R P G++
Sbjct: 230 YPSVFPPFVLFEEPQESQ--------------PSPAPMPAPIRSYRDPLDEEEAPHGANG 367
Query: 121 RKSRGQLPEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTIN 180
S + N VD S+ K + + + + K D+ P
Sbjct: 368 ETSSPMEGLNGNSVDNSQKRAVPPMHSCKYSQKRSK-KTKESQFDLSPSV---------- 514
Query: 181 PGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQI-DEKAKGSHSGAKRPDLKAGTLMNTK 239
G + + DG R+ V LM Y+ +RR+L Q+ D K + KR DL+A M K
Sbjct: 515 -GGISVATREDGDRELVNLVLMTYDSLRRRLCQLEDAKELNTTMAIKRADLRASNAMTVK 691
Query: 240 GIRANSRKRIGVVPGVEVGDIFFFRFELCLVGL 272
R N+R+R+G VPGVE+GDIFF R E+CLVGL
Sbjct: 692 AFRTNTRRRVGAVPGVEIGDIFFLRMEMCLVGL 790
>TC218322 similar to UP|SUV9_ARATH (Q9T0G7) Probable histone-lysine
N-methyltransferase, H3 lysine-9 specific 9 (Histone
H3-K9 methyltransferase 9) (H3-K9-HMTase 9) (Suppressor
of variegation 3-9 homolog 9) (Su(var)3-9 homolog 9) ,
partial (26%)
Length = 810
Score = 109 bits (272), Expect = 5e-24
Identities = 66/158 (41%), Positives = 94/158 (58%), Gaps = 8/158 (5%)
Frame = +1
Query: 522 PSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVI--DNARVEELSG 579
P CRNRV+Q GLK RLEVFR++ GWG+RS D I+AG FICEY G V+ + AR+ ++G
Sbjct: 1 PHCRNRVTQKGLKNRLEVFRSRETGWGVRSMDLIQAGAFICEYTGVVLTREQARLLTMNG 180
Query: 580 EN--EDDYIFDSTRIYQQLEVFSSDVEAPKIPS--PL--YITARNEGNVARFMNHSCTPN 633
++ + D + L + S+ P PS PL + NVA +M+HS TPN
Sbjct: 181 DSLIYPNRFTDRWAEWGDLSMIDSNFVRPSYPSIPPLDFAMDVSRMRNVACYMSHSSTPN 360
Query: 634 VLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGI 671
VL + V+ ++ N + +A+ IPPM EL+ DYG+
Sbjct: 361 VLVQFVLYDHNNLMFPRLMLFAMESIPPMRELSLDYGV 474
>BF010305 weakly similar to GP|13517745|gb| SUVH2 {Arabidopsis thaliana},
partial (12%)
Length = 418
Score = 92.4 bits (228), Expect = 6e-19
Identities = 44/87 (50%), Positives = 52/87 (59%)
Frame = +3
Query: 467 VESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRN 526
V + GC C GC G C C KNGG PY+ G L K +++ECGP C CPP CRN
Sbjct: 165 VGKATGCDCVDGCGDG---CFCAMKNGGEFPYTLQGHLVRGKPLIFECGPFCSCPPHCRN 335
Query: 527 RVSQGGLKLRLEVFRTKGKGWGLRSWD 553
RV+ GLK LEVFR+K G+RS D
Sbjct: 336 RVAHNGLKYMLEVFRSKQTSRGVRSLD 416
>TC229925 similar to UP|Q946J2 (Q946J2) Suppressor of variegation related 1,
partial (22%)
Length = 935
Score = 73.2 bits (178), Expect(2) = 9e-16
Identities = 54/156 (34%), Positives = 74/156 (46%), Gaps = 7/156 (4%)
Frame = +1
Query: 544 GKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDV 603
GKGWGLR+ + + G F+CE+ GE++ ++EL E Y + Y L
Sbjct: 142 GKGWGLRTLEDLPKGAFVCEFVGEIL---TLKELH-ERNLKYPKNGKYTYPILLDADWGS 309
Query: 604 EAPKIPSPLYITARNEGNVARFMNHSC-TPNVLWRPVVRENKNEADLHVAFYAIRHIPPM 662
K L + A + GN ARF+NH C N++ PV E H AF+ R I
Sbjct: 310 GIVKDREALCLYAASYGNAARFINHRCLDANLIEIPVEVEGPTHHYYHFAFFTSRKIAAQ 489
Query: 663 MELTYDYGIVL------PLKVGQKKKKCLCGSVKCR 692
ELT+DYGI P+++ Q C CGS CR
Sbjct: 490 EELTWDYGINFDDHDDHPVELFQ----CRCGSKFCR 585
Score = 28.9 bits (63), Expect(2) = 9e-16
Identities = 11/32 (34%), Positives = 17/32 (52%)
Frame = +2
Query: 508 KSVVYECGPSCHCPPSCRNRVSQGGLKLRLEV 539
+ + EC C C C NRV Q G+ +L++
Sbjct: 47 RKFIKECWSKCGCGKHCGNRVVQRGITCKLQM 142
>TC229811 similar to UP|Q9FIH7 (Q9FIH7) Similarity to SET-domain
transcriptional regulator, partial (11%)
Length = 1108
Score = 78.2 bits (191), Expect = 1e-14
Identities = 53/160 (33%), Positives = 79/160 (49%)
Frame = +1
Query: 534 KLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIY 593
K L R+K WGL + +PI A F+ EY GE+I R+ ++ R Y
Sbjct: 88 KKHLRFQRSKIHDWGLLALEPIEAEDFVIEYIGELI-RPRISDI-----------RERQY 231
Query: 594 QQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAF 653
+++ + SS + ++ + A G +ARF+NHSC PN + + E + + +
Sbjct: 232 EKMGIGSSYLF--RLDDGYVVDATKRGGIARFVNHSCEPNCYTKVISVEGQKK----IFI 393
Query: 654 YAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRG 693
YA RHI E+TY+Y L +KK C CGS KCRG
Sbjct: 394 YAKRHIAAGEEITYNYKFPLE----EKKIPCNCGSRKCRG 501
>TC222698 similar to UP|SUV8_ARATH (Q9C5P0) Histone-lysine
N-methyltransferase, H3 lysine-9 specific 8 (Histone
H3-K9 methyltransferase 8) (H3-K9-HMTase 8) (Suppressor
of variegation 3-9 homolog 8) (Su(var)3-9 homolog 8) ,
partial (7%)
Length = 523
Score = 75.1 bits (183), Expect = 1e-13
Identities = 35/66 (53%), Positives = 43/66 (65%), Gaps = 3/66 (4%)
Frame = +3
Query: 633 NVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGI---VLPLKVGQKKKKCLCGSV 689
NV W+PV N++ LH+AF+A+RHIPPM ELTYDYG + +KKC CGS
Sbjct: 3 NVFWQPVXYAENNQSFLHIAFFALRHIPPMTELTYDYGCSGHADGSSAPKGRKKCSCGSS 182
Query: 690 KCRGYF 695
KCRG F
Sbjct: 183 KCRGSF 200
>AW306371 similar to GP|13517751|gb| SUVH5 {Arabidopsis thaliana}, partial
(7%)
Length = 430
Score = 63.5 bits (153), Expect = 3e-10
Identities = 32/72 (44%), Positives = 46/72 (63%)
Frame = +2
Query: 332 KLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFK 391
+L+RGNLAL+ S N VRVIRG + K YVYDGLY +++ W ++ G V++
Sbjct: 5 ELKRGNLALKNSSEEKNPVRVIRGS-ESMDDKYKTYVYDGLYVVESYWQDRGSHGKLVYR 181
Query: 392 YKLVRLPGQPQA 403
++L R+PGQ A
Sbjct: 182 FRLKRIPGQKLA 217
>TC232298 similar to GB|AAO64916.1|29029074|BT005981 At5g53430 {Arabidopsis
thaliana;} , partial (16%)
Length = 768
Score = 62.0 bits (149), Expect = 9e-10
Identities = 44/153 (28%), Positives = 67/153 (43%), Gaps = 4/153 (2%)
Frame = +1
Query: 546 GWGLRSWDPIRAGTFICEYAG----EVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSS 601
GWGL + I+ G + EY G + + R E+ E +D Y+F
Sbjct: 325 GWGLFARRDIQEGEMVVEYRGVHVRRSVADLREEKYRSEGKDCYLF-------------- 462
Query: 602 DVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPP 661
KI + + A N GN+AR +NHSC PN R + + + + A ++
Sbjct: 463 -----KISEEVVVDATNRGNIARLINHSCMPNCYARIM---SLGDQGSRIVLIAKTNVSA 618
Query: 662 MMELTYDYGIVLPLKVGQKKKKCLCGSVKCRGY 694
ELTYDY + P + + K CLC + CR +
Sbjct: 619 GEELTYDY-LFDPDERDELKVPCLCKAPNCRRF 714
>TC229209 similar to UP|Q9C5X4 (Q9C5X4) Trithorax-like protein 1, partial
(28%)
Length = 1537
Score = 61.2 bits (147), Expect = 1e-09
Identities = 45/158 (28%), Positives = 71/158 (44%)
Frame = +2
Query: 536 RLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQ 595
RL +++ G+G+ + P + G + EY GE+ V + + +I++S
Sbjct: 659 RLAFGKSRIHGFGIFAKHPYKGGDMVIEYTGEL-----VRPPIADRREHFIYNS------ 805
Query: 596 LEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYA 655
+ + +I I A G++A +NHSC PN R V+ N +E H+ +A
Sbjct: 806 --LVGAGTYMFRIDDERVIDATRAGSIAHLINHSCAPNCYSR-VISVNGDE---HIIIFA 967
Query: 656 IRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRG 693
R I ELTYDY ++ C CG KCRG
Sbjct: 968 KRDIKQWEELTYDYRFFSI----DERLACYCGFPKCRG 1069
>CD396083
Length = 227
Score = 42.7 bits (99), Expect(2) = 2e-08
Identities = 18/20 (90%), Positives = 19/20 (95%)
Frame = -1
Query: 665 LTYDYGIVLPLKVGQKKKKC 684
LTYDYG VLPLKVGQ+KKKC
Sbjct: 218 LTYDYGTVLPLKVGQRKKKC 159
Score = 34.7 bits (78), Expect(2) = 2e-08
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = -2
Query: 681 KKKCLCGSVKCRGYFC 696
K+ LCGSVKC+GYFC
Sbjct: 169 KRNALCGSVKCKGYFC 122
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,846,496
Number of Sequences: 63676
Number of extensions: 506548
Number of successful extensions: 3393
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 3265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3350
length of query: 696
length of database: 12,639,632
effective HSP length: 104
effective length of query: 592
effective length of database: 6,017,328
effective search space: 3562258176
effective search space used: 3562258176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0098b.1


