

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098a.6
(626 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
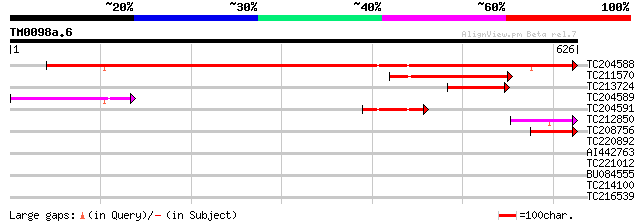
Score E
Sequences producing significant alignments: (bits) Value
TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 803 0.0
TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 165 5e-41
TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-... 112 4e-25
TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosida... 103 3e-22
TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydr... 92 9e-19
TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofur... 58 1e-08
TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosida... 58 1e-08
TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-rel... 35 0.077
AI442763 30 4.2
TC221012 29 7.2
BU084555 29 7.2
TC214100 similar to GB|AAC03224.1|603365|SCE9781 Yer126cp {Sacch... 29 7.2
TC216539 homologue to UP|PSA6_SOYBN (O48551) Proteasome subunit ... 28 9.4
>TC204588 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(84%)
Length = 2003
Score = 803 bits (2073), Expect = 0.0
Identities = 382/594 (64%), Positives = 469/594 (78%), Gaps = 8/594 (1%)
Frame = +2
Query: 41 GTGGLWAELVNNRGFEAGGTQTPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRMDVL 100
GT GLWAELV+NRGFEAGG TPSNI PW+I+G ESL+ ++T+ +SCF+RNKVALR++VL
Sbjct: 8 GTRGLWAELVSNRGFEAGGPNTPSNIDPWSIIGNESLINIETDRTSCFDRNKVALRLEVL 187
Query: 101 CDQ-----CPSDGVGVSNPGYWGMNVVQKKEYKVVFFVKSTGSLDMTISFKKAEDGGILA 155
CD CP+DGVGV NPG+WGMN+ Q K+YKVVF+V+ST SL++T+S + G LA
Sbjct: 188 CDSKGDNICPADGVGVYNPGFWGMNIEQGKKYKVVFYVRSTESLNLTVSLTGSNGVGNLA 367
Query: 156 SQNVKASASEVANWKRMELKLVPTASNPKATLHLTTTQKGVIWLDQVSAMPVDTFKGHGF 215
S + SA + +NW ++E LV A++ + L LTTT KGVIWLDQVSAMP+DT+KGHGF
Sbjct: 368 SSVITGSALDFSNWTKVEKILVAKATDHNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGF 547
Query: 216 RTDLVNMVLELKPAFIRFPGGCFIEGQTLRNAFRWKDSVGPWEERPGHFGDVWGSWTDEG 275
RTDLV M+ LKP FIRFPGGCF+EG+ LRNAFRWK S+GPWEERPGHFGDVW WTD+G
Sbjct: 548 RTDLVEMLTALKPRFIRFPGGCFVEGEWLRNAFRWKTSIGPWEERPGHFGDVWMYWTDDG 727
Query: 276 LGYFEGLQLAEDIGAKPIWVFNNGISHTDQIDTSVITPFVQEALDGIEFARGPATSKWGS 335
LGY+E LQLAED+G PIWVFNNG+SH DQ+DTS + P VQEALDG+EFARG TSKWGS
Sbjct: 728 LGYYEFLQLAEDLGTAPIWVFNNGVSHNDQVDTSAVLPLVQEALDGLEFARGDRTSKWGS 907
Query: 336 LRKSMGHPEPFDLKYVAIGNEDCGKKNYLGNYLAFYNAIRKAYPDIQMISNCDASSKPLD 395
LR +MGHP PFDL+YVA+GNEDCGKKNY GNYL FY+AI+ AYPDIQ+ISNCD SS+PLD
Sbjct: 908 LRAAMGHPAPFDLRYVAVGNEDCGKKNYRGNYLKFYSAIKSAYPDIQIISNCDGSSRPLD 1087
Query: 396 HPADLYDYHTYPNNPSNMFNNAHVFDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGVSEA 455
HPAD+YDYH Y N ++MF+ A+ FD T R GPKAFVSEYA+ G + A G+L+ ++EA
Sbjct: 1088HPADMYDYHVY-TNANDMFSRANAFDHTSRNGPKAFVSEYAVTG-KDAGTGSLLAALAEA 1261
Query: 456 GFLIGLEKNSDHVAMAAYAPLFVNADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGA 515
GFLIGLEKNSD + MA+YAPLFVNA+DR+WNPDAIVFNS Q+YGTPSYWV F ES+GA
Sbjct: 1262GFLIGLEKNSDVIQMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWVQLFFSESSGA 1441
Query: 516 TFLASTLQTPDPSSFDASTILWQNPQDKKTYLKIKVANLGNNQVKLGIVVHGLESSKII- 574
T L+++LQ P +S AS I +QN DKK Y++IKV N G V L I + GLE + +
Sbjct: 1442TLLSTSLQAPSSNSLVASAITFQNSVDKKNYIRIKVVNFGTTAVNLKISLDGLEPNSLQL 1621
Query: 575 --GTKTVLTSKNALDENTFLEPRKIVPQQTPLEEASANMNVELPPLSVTSFDIL 626
TKTVLTS N +DEN+F +P+K+VP Q+ L+ +MNV +PP S TSFD++
Sbjct: 1622SGSTKTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKDMNVTVPPRSFTSFDLV 1783
>TC211570 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(20%)
Length = 414
Score = 165 bits (418), Expect = 5e-41
Identities = 82/136 (60%), Positives = 98/136 (71%)
Frame = +3
Query: 420 FDKTPRKGPKAFVSEYALVGDQQAKLGTLIGGVSEAGFLIGLEKNSDHVAMAAYAPLFVN 479
FD T R GPKAFVSEYA+ D A G L+ V+EA FLIGLEKNSD M +YAPLF+N
Sbjct: 6 FDNTSRSGPKAFVSEYAVKSD--AANGNLLAAVAEAAFLIGLEKNSDIGEMVSYAPLFLN 179
Query: 480 ADDRKWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQN 539
+D++W PDAIVFNS QVYGTPSYWV +F ES+GATF+ STL T + AS I+WQ+
Sbjct: 180 INDKRWIPDAIVFNSYQVYGTPSYWVQKLFIESSGATFIDSTLSTTSSNKLAASVIIWQD 359
Query: 540 PQDKKTYLKIKVANLG 555
DK+ YL+IK N G
Sbjct: 360 SSDKRNYLRIKAVNFG 407
>TC213724 weakly similar to UP|Q944F3 (Q944F3) Arabinosidase ARA-1, partial
(10%)
Length = 595
Score = 112 bits (281), Expect = 4e-25
Identities = 54/68 (79%), Positives = 58/68 (84%)
Frame = +3
Query: 484 KWNPDAIVFNSNQVYGTPSYWVTHMFKESNGATFLASTLQTPDPSSFDASTILWQNPQDK 543
+W+PDAIVFNSNQVY TPSY V MF+ESNGATFL S LQTPDP S DAS ILW+N QDK
Sbjct: 141 RWSPDAIVFNSNQVYETPSYRVQFMFRESNGATFLKSQLQTPDPDSVDASAILWKNLQDK 320
Query: 544 KTYLKIKV 551
KTYLKIKV
Sbjct: 321 KTYLKIKV 344
>TC204589 similar to UP|Q76LU4 (Q76LU4) Alpha-L-arabinofuranosidase, partial
(14%)
Length = 808
Score = 103 bits (256), Expect = 3e-22
Identities = 63/144 (43%), Positives = 82/144 (56%), Gaps = 5/144 (3%)
Frame = +3
Query: 1 CYVADAAGDQTSTLTVDLKSA-GRPIPETLFGVFYEEINHAGTGGLWAELVNNRGFEAGG 59
C+ D T+TLTV+ A G+PIPETLFG+F+EEINHAG GGLWAELV+NRGFEAGG
Sbjct: 294 CFATKKHADLTTTLTVNASDASGKPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGG 473
Query: 60 TQTPSNIAPWTIVGKESLVLLQTELSSCFERNKVALRMDVLCDQ----CPSDGVGVSNPG 115
TPSNIA ++ +L+ R +V + Q P+DGV V+
Sbjct: 474 PNTPSNIALGQLLEMNH**ILKLIAPLALTRTRVHSDWRCM*HQX*HIWPADGV-VAITL 650
Query: 116 YWGMNVVQKKEYKVVFFVKSTGSL 139
GMN+ Q +E F+ + L
Sbjct: 651 VLGMNIEQXEEXXXGFYCRQLDHL 722
>TC204591 homologue to UP|Q6RIB6 (Q6RIB6) Cytosolic malate dehydrogenase ,
partial (84%)
Length = 1604
Score = 91.7 bits (226), Expect = 9e-19
Identities = 44/73 (60%), Positives = 57/73 (77%)
Frame = +2
Query: 390 SSKPLDHPADLYDYHTYPNNPSNMFNNAHVFDKTPRKGPKAFVSEYALVGDQQAKLGTLI 449
+S+PLDHPAD+YDYH Y N ++MF+ A+ FD T R GPKAFVSEYA+ G + A G+L+
Sbjct: 1391 ASRPLDHPADMYDYHVY-TNANDMFSRANAFDHTSRNGPKAFVSEYAVTG-KDAGTGSLL 1564
Query: 450 GGVSEAGFLIGLE 462
++EAGFLIGLE
Sbjct: 1565 AALAEAGFLIGLE 1603
>TC212850 weakly similar to UP|Q76LU3 (Q76LU3) Alpha-L-arabinofuranosidase,
partial (3%)
Length = 558
Score = 57.8 bits (138), Expect = 1e-08
Identities = 38/109 (34%), Positives = 54/109 (48%), Gaps = 36/109 (33%)
Frame = +2
Query: 554 LGNNQVKLGIVVHGLESSKII-GTKTVLTSKNALDENTFLEP------------------ 594
+G+NQ+ I ++G ESS TKTVLTSKNALDEN+ P
Sbjct: 8 VGSNQINFKISLNGFESSNATKATKTVLTSKNALDENSLANPTNVHITSFILFLLL*TNN 187
Query: 595 -----------------RKIVPQQTPLEEASANMNVELPPLSVTSFDIL 626
+IVP+++PL+ A+ +N LPP+S+T FD+L
Sbjct: 188 TYNN*LS*HLMFICLFCTQIVPKRSPLQSANNEINDILPPVSLTVFDLL 334
>TC208756 similar to UP|Q7X9G7 (Q7X9G7) Alpha-L-arabinofuranosidase, partial
(5%)
Length = 544
Score = 57.8 bits (138), Expect = 1e-08
Identities = 29/51 (56%), Positives = 36/51 (69%)
Frame = +1
Query: 576 TKTVLTSKNALDENTFLEPRKIVPQQTPLEEASANMNVELPPLSVTSFDIL 626
T TVLTS N +DEN+FLEP+K+VPQ +PLE ++ V LP SV S D L
Sbjct: 28 TITVLTSTNVMDENSFLEPKKVVPQTSPLEIDDKHLIVLLPSYSVISLDFL 180
>TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-related
protein, partial (5%)
Length = 647
Score = 35.4 bits (80), Expect = 0.077
Identities = 15/37 (40%), Positives = 27/37 (72%)
Frame = +3
Query: 120 NVVQKKEYKVVFFVKSTGSLDMTISFKKAEDGGILAS 156
N+ + ++YKVVF+ K+ G++++ +SF +E G LAS
Sbjct: 129 NIEKGQKYKVVFYAKADGAINLDVSFVGSEKGEKLAS 239
>AI442763
Length = 426
Score = 29.6 bits (65), Expect = 4.2
Identities = 14/32 (43%), Positives = 18/32 (55%), Gaps = 1/32 (3%)
Frame = -2
Query: 266 DVWG-SWTDEGLGYFEGLQLAEDIGAKPIWVF 296
++WG SW LGYF + L +D K WVF
Sbjct: 131 EIWGFSWVYLTLGYFYFVPLIDDFFLKRTWVF 36
>TC221012
Length = 436
Score = 28.9 bits (63), Expect = 7.2
Identities = 19/57 (33%), Positives = 32/57 (55%), Gaps = 5/57 (8%)
Frame = -1
Query: 538 QNPQDKKTYLKIKVANLGNNQV-----KLGIVVHGLESSKIIGTKTVLTSKNALDEN 589
+NPQ +KT LKI++ + NQ+ L I +H L I ++ +L+ KN + E+
Sbjct: 271 RNPQ*QKTQLKIELTMINPNQI*IM*D*LRIRIH*LRYMFICTSEKLLSKKNKVAES 101
>BU084555
Length = 421
Score = 28.9 bits (63), Expect = 7.2
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = -2
Query: 485 WNPDAIVFNSNQVYGTPSYWV 505
WNPDA + N +Y T +WV
Sbjct: 345 WNPDATLNKQNSIYTTIVFWV 283
>TC214100 similar to GB|AAC03224.1|603365|SCE9781 Yer126cp {Saccharomyces
cerevisiae;} , partial (64%)
Length = 1072
Score = 28.9 bits (63), Expect = 7.2
Identities = 10/21 (47%), Positives = 13/21 (61%)
Frame = -1
Query: 485 WNPDAIVFNSNQVYGTPSYWV 505
WNPDA + N +Y T +WV
Sbjct: 883 WNPDATLNKQNSIYTTIVFWV 821
>TC216539 homologue to UP|PSA6_SOYBN (O48551) Proteasome subunit alpha type 6
(20S proteasome alpha subunit A) (20S proteasome
subunit alpha-1) (Proteasome iota subunit) , complete
Length = 1073
Score = 28.5 bits (62), Expect = 9.4
Identities = 13/31 (41%), Positives = 19/31 (60%), Gaps = 2/31 (6%)
Frame = -3
Query: 379 PDIQMISNCDASSKPLDH--PADLYDYHTYP 407
P IQ + CD S+ LDH A+++ +H YP
Sbjct: 675 PSIQHLLPCDQSNDQLDHICKAEVHIHHQYP 583
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,527,200
Number of Sequences: 63676
Number of extensions: 384624
Number of successful extensions: 1652
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1638
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1644
length of query: 626
length of database: 12,639,632
effective HSP length: 103
effective length of query: 523
effective length of database: 6,081,004
effective search space: 3180365092
effective search space used: 3180365092
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0098a.6


