

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0097b.12
(292 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
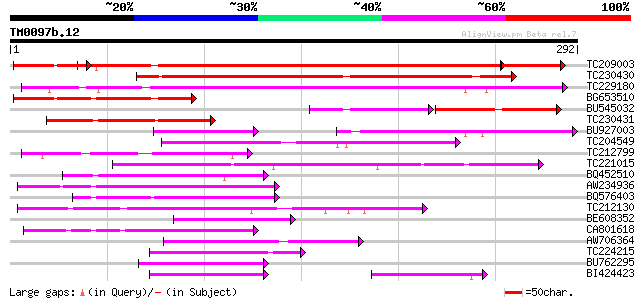
Score E
Sequences producing significant alignments: (bits) Value
TC209003 weakly similar to GB|AAP13422.1|30023778|BT006314 At2g0... 343 e-106
TC230430 weakly similar to UP|Q9FHQ4 (Q9FHQ4) Similarity to secr... 206 1e-53
TC229180 similar to UP|P93806 (P93806) F19P19.1, partial (84%) 149 1e-36
BG653510 similar to GP|9757976|dbj| contains similarity to secre... 122 3e-28
BU545032 70 6e-24
TC230431 weakly similar to UP|Q9FHQ4 (Q9FHQ4) Similarity to secr... 92 3e-19
BU927003 75 3e-14
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 73 2e-13
TC212799 weakly similar to UP|Q9FNC8 (Q9FNC8) Gb|AAB80651.1, par... 70 1e-12
TC221015 similar to UP|Q9FNE1 (Q9FNE1) Receptor-like serine/thre... 66 2e-11
BQ452510 similar to GP|6633857|gb|A F2K11.7 {Arabidopsis thalian... 66 2e-11
AW234936 65 3e-11
BQ576403 similar to PIR|D85065|D850 receptor protein kinase-like... 63 1e-10
TC212130 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 57 1e-08
BE608352 similar to PIR|F86177|F861 protein F19P19.1 [imported] ... 55 3e-08
CA801618 55 4e-08
AW706364 similar to GP|8777384|dbj| 33 kDa secretory protein-lik... 52 4e-07
TC224215 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like prot... 51 7e-07
BU762295 weakly similar to PIR|D85065|D850 receptor protein kina... 49 3e-06
BI424423 weakly similar to GP|4530126|gb|A receptor-like protein... 48 5e-06
>TC209003 weakly similar to GB|AAP13422.1|30023778|BT006314 At2g01660
{Arabidopsis thaliana;} , partial (48%)
Length = 1383
Score = 343 bits (880), Expect(2) = e-106
Identities = 168/232 (72%), Positives = 192/232 (82%), Gaps = 12/232 (5%)
Frame = +1
Query: 36 PKNTPVSD-----------PVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGL 84
P++TP S P+YE VNSLLTSLVNSA+F NYNNFTVPA S DT+YGL
Sbjct: 199 PRSTPSSSAAAPRPSSRRVPLYENTVNSLLTSLVNSAAFANYNNFTVPA---SSDTVYGL 369
Query: 85 YQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVEDKTMVVK 144
+QCRGDL NDQC RCV+RAV+QLGTLC A GGALQL+GCFVKYDN F+GVEDKT+V K
Sbjct: 370 FQCRGDLPNDQCSRCVSRAVTQLGTLCFASCGGALQLDGCFVKYDNTTFIGVEDKTLVSK 549
Query: 145 KCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCL 204
KCGPS+GLTSDAL+RR+AVLAYLQTSDGVY+T+R S YGDFQG AQCTGDLSPS+CQDCL
Sbjct: 550 KCGPSVGLTSDALSRREAVLAYLQTSDGVYKTFRTSGYGDFQGVAQCTGDLSPSQCQDCL 729
Query: 205 SESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR-HSDDDSNHNDDEIEK 255
S+SIQR KTECG + W E+YLAKCYARYSE G SR +++DDSNHNDDEI +
Sbjct: 730 SDSIQRFKTECGPTNWAEIYLAKCYARYSEAGTRSRANNNDDSNHNDDEIRE 885
Score = 61.2 bits (147), Expect(2) = e-106
Identities = 31/33 (93%), Positives = 32/33 (96%)
Frame = +2
Query: 254 EKTLAILIGLIAGVALIIVFLSFLSKVCEKQRG 286
EKTLAILIGLIAGVALIIVFLSFLSKVCEK +G
Sbjct: 881 EKTLAILIGLIAGVALIIVFLSFLSKVCEKHKG 979
Score = 48.9 bits (115), Expect = 3e-06
Identities = 27/41 (65%), Positives = 34/41 (82%), Gaps = 1/41 (2%)
Frame = +3
Query: 3 LLLRLLFILTL-SAITTPSTSSSIDTFIFGGCSQPKNTPVS 42
++L LLF++ + SA+TTPSTS+ IDTFIFGGCSQ K TP S
Sbjct: 138 MVLVLLFLMAIFSAMTTPSTSA-IDTFIFGGCSQAKFTPGS 257
>TC230430 weakly similar to UP|Q9FHQ4 (Q9FHQ4) Similarity to secretory
protein, partial (55%)
Length = 659
Score = 206 bits (524), Expect = 1e-53
Identities = 100/196 (51%), Positives = 130/196 (66%)
Frame = +3
Query: 66 YNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
YNNFTV + D +YG YQCRGDL C CVARAV++ G +C GG++QL+GCF
Sbjct: 9 YNNFTVVGSTQQ-DAVYGXYQCRGDLAMPDCAACVARAVTRAGDICRGTCGGSVQLDGCF 185
Query: 126 VKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDF 185
VKYDN FLG +DK +V+KKCGPS+G DA+ RDAVLA L + G +R+ G
Sbjct: 186 VKYDNATFLGAQDKAVVLKKCGPSVGYNPDAMGSRDAVLAGLAAAGG---NFRVGGSGGV 356
Query: 186 QGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSRHSDDD 245
G AQCTGDLS ECQDC++E+I RLK++CG + +G+M+L KCYARYS GG H D+
Sbjct: 357 HGVAQCTGDLSYGECQDCVAEAISRLKSDCGTADYGDMFLGKCYARYSVGGAH----DES 524
Query: 246 SNHNDDEIEKTLAILI 261
H + + L +L+
Sbjct: 525 KAHGKAQHRRMLPLLL 572
>TC229180 similar to UP|P93806 (P93806) F19P19.1, partial (84%)
Length = 1200
Score = 149 bits (377), Expect = 1e-36
Identities = 92/296 (31%), Positives = 148/296 (49%), Gaps = 15/296 (5%)
Frame = +2
Query: 7 LLFILTLSAITTP-----STSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLVN 59
+L +L ++++ P STSS T ++ GCS+ P +DP VY +++L SLV+
Sbjct: 269 VLLVLFITSLLEPPHVAESTSSDYSTLVYKGCSKE---PFTDPNGVYSQALSALFGSLVS 439
Query: 60 SASFTNYNNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
++ + T+GSG +T+ GL+QCRGDL N CY CV+R LC
Sbjct: 440 QSTKAKFYK----TTSGSGQNTITGLFQCRGDLTNSDCYNCVSRLPVLCDKLCGKTTAAR 607
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWR 178
+QL GC++ Y+ F + M+ K CG + RRD + ++ +
Sbjct: 608 VQLLGCYILYEVAGFSQISGMQMLYKTCGATNAAGRGFEERRDTAFSVMENGVVSGHGFY 787
Query: 179 ISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS--EGG 236
++Y QC GD+ S+C +C+ ++QR + ECG+S G+++L KC+ YS G
Sbjct: 788 TTSYQSLYVMGQCEGDVGDSDCGECVKNAVQRAQVECGSSISGQVFLHKCFISYSYYPNG 967
Query: 237 DHSRHSDD-----DSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQRGM 287
SR S S+ + KT AI++G A VA +++ L F + +K GM
Sbjct: 968 VPSRSSSSSSASYSSSSSGQNPGKTAAIILGGAAAVAFLVILLLFARSLKKKHDGM 1135
>BG653510 similar to GP|9757976|dbj| contains similarity to secretory
protein~gene_id:K12B20.12 {Arabidopsis thaliana},
partial (24%)
Length = 323
Score = 122 bits (305), Expect = 3e-28
Identities = 68/95 (71%), Positives = 76/95 (79%), Gaps = 1/95 (1%)
Frame = +1
Query: 3 LLLRLLFILTL-SAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSA 61
L L LLF++ + SA+TTPSTS+ IDTFIFGGCSQ K TP S YE VNSLLTSLVNSA
Sbjct: 55 LSLSLLFLMAIFSAMTTPSTSA-IDTFIFGGCSQAKFTPGS--AYENTVNSLLTSLVNSA 225
Query: 62 SFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQC 96
+F NYNNFTVPA S DT+YGL+QCRGDL NDQC
Sbjct: 226 AFANYNNFTVPA---SSDTVYGLFQCRGDLPNDQC 321
>BU545032
Length = 642
Score = 69.7 bits (169), Expect(2) = 6e-24
Identities = 33/66 (50%), Positives = 47/66 (71%), Gaps = 1/66 (1%)
Frame = -1
Query: 220 WGEMYLAKCYARYSEGGDHSRH-SDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLS 278
+G+M+L KCYARYS GG H + D SNH KT +I+IG +AGVA++I+F +F+S
Sbjct: 450 YGDMFLGKCYARYSVGGAHDESKAHDKSNHGG---VKTFSIIIGSLAGVAILIIFFAFMS 280
Query: 279 KVCEKQ 284
K+C +Q
Sbjct: 279 KICGRQ 262
Score = 58.5 bits (140), Expect(2) = 6e-24
Identities = 29/64 (45%), Positives = 38/64 (59%)
Frame = -3
Query: 155 DALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTE 214
DA+ RDAV A L + G +R+ G G AQCTG ECQDC++E+I RLK++
Sbjct: 637 DAMGSRDAVXAGLAAAGG---NFRVGGSGGVHGVAQCTGXXXXGECQDCVAEAISRLKSD 467
Query: 215 CGAS 218
C S
Sbjct: 466 CXXS 455
Score = 32.3 bits (72), Expect = 0.26
Identities = 16/44 (36%), Positives = 24/44 (54%)
Frame = -3
Query: 68 NFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLC 111
NF V + G ++G+ QC G +C CVA A+S+L + C
Sbjct: 583 NFRVGGSGG----VHGVAQCTGXXXXGECQDCVAEAISRLKSDC 464
>TC230431 weakly similar to UP|Q9FHQ4 (Q9FHQ4) Similarity to secretory
protein, partial (29%)
Length = 396
Score = 92.0 bits (227), Expect = 3e-19
Identities = 48/87 (55%), Positives = 60/87 (68%)
Frame = +3
Query: 20 STSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGD 79
S SS TF++ GC+Q + TP S YE +NSLLTSLVNSA+++ YNNFTV + D
Sbjct: 141 SECSSTSTFLYSGCTQQRYTPNSP--YEWNINSLLTSLVNSATYSAYNNFTVVGST-QQD 311
Query: 80 TLYGLYQCRGDLNNDQCYRCVARAVSQ 106
+YGLYQCRGDL C CVARAV++
Sbjct: 312 AVYGLYQCRGDLAMPDCAACVARAVTR 392
>BU927003
Length = 446
Score = 75.5 bits (184), Expect = 3e-14
Identities = 44/136 (32%), Positives = 67/136 (48%), Gaps = 12/136 (8%)
Frame = +2
Query: 169 TSDGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKC 228
T G Y T +Y QC GD+ S+C +C+ ++QR + ECG+S G++YL KC
Sbjct: 29 TGHGFYAT----SYQSLYVMGQCEGDVGDSDCGECVKSAVQRAQVECGSSISGQVYLHKC 196
Query: 229 YARYS--EGGDHSRHS----------DDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSF 276
+ YS G RHS S+ + KT+AI++G AGVA +++ L F
Sbjct: 197 FISYSYYPNGVPGRHSSSASSSSNSYSSSSSSSGQNTGKTVAIIVGGAAGVAFLVICLMF 376
Query: 277 LSKVCEKQRGMVLWYM 292
+ +K+ W M
Sbjct: 377 ARSLKKKRDDY*QWKM 424
Score = 41.6 bits (96), Expect = 4e-04
Identities = 19/54 (35%), Positives = 27/54 (49%)
Frame = +2
Query: 75 AGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
A S +LY + QC GD+ + C CV AV + C + G + L CF+ Y
Sbjct: 47 ATSYQSLYVMGQCEGDVGDSDCGECVKSAVQRAQVECGSSISGQVYLHKCFISY 208
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 72.8 bits (177), Expect = 2e-13
Identities = 49/171 (28%), Positives = 72/171 (41%), Gaps = 17/171 (9%)
Frame = +3
Query: 79 DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVED 138
D +YGL+ CRGD+ + C +CV A +L + CS + + + C V+Y N F D
Sbjct: 27 DRVYGLFMCRGDVPSALCQQCVVNATGRLRSQCSLAKQAVIWYDECTVRYSNRSFFSTVD 206
Query: 139 KTMVVKKCGPSIGLTSDA-LTRRDAVLAYL-----QTSDG-----------VYRTWRISA 181
P +GL + A ++ +D+ + L +T+D IS
Sbjct: 207 TR-------PRVGLLNTANISNQDSFMRLLFQTINRTADEAANFSVGLKKYAVNQANISG 365
Query: 182 YGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
+ AQCT DLS C+ CLS I L C G + C RY
Sbjct: 366 FQSLYCLAQCTPDLSQENCRSCLSGVIGDLPWCCQGKQGGRVLYPSCNVRY 518
Score = 39.7 bits (91), Expect = 0.002
Identities = 18/57 (31%), Positives = 28/57 (48%)
Frame = +3
Query: 80 TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGV 136
+LY L QC DL+ + C C++ + L C +GG + C V+Y+ F V
Sbjct: 372 SLYCLAQCTPDLSQENCRSCLSGVIGDLPWCCQGKQGGRVLYPSCNVRYELYPFYRV 542
Score = 36.2 bits (82), Expect = 0.018
Identities = 14/47 (29%), Positives = 25/47 (52%)
Frame = +3
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G C GD+ + CQ C+ + RL+++C + ++ +C RYS
Sbjct: 39 GLFMCRGDVPSALCQQCVVNATGRLRSQCSLAKQAVIWYDECTVRYS 179
>TC212799 weakly similar to UP|Q9FNC8 (Q9FNC8) Gb|AAB80651.1, partial (11%)
Length = 430
Score = 70.1 bits (170), Expect = 1e-12
Identities = 43/124 (34%), Positives = 64/124 (50%), Gaps = 5/124 (4%)
Frame = +2
Query: 7 LLFILTLSA---ITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASF 63
LLF L++S + TP T+S I+ GC+ K Y + LL SLV++++
Sbjct: 92 LLFFLSISFPLFLLTPITTSDNTNLIYKGCADQK----MQGQYSQNLKPLLDSLVSASAQ 259
Query: 64 TNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSA--VRGGALQL 121
+ A + + L G YQCRGDL+N +CY CV++ + LG LC V +QL
Sbjct: 260 KGF-------AATTQNALTGAYQCRGDLSNSECYNCVSKIPNMLGRLCGGDDVAAARVQL 418
Query: 122 EGCF 125
GC+
Sbjct: 419 SGCY 430
Score = 36.2 bits (82), Expect = 0.018
Identities = 19/45 (42%), Positives = 24/45 (53%), Gaps = 2/45 (4%)
Frame = +2
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGAS--TWGEMYLAKCY 229
GA QC GDLS SEC +C+S+ L CG + L+ CY
Sbjct: 296 GAYQCRGDLSNSECYNCVSKIPNMLGRLCGGDDVAAARVQLSGCY 430
>TC221015 similar to UP|Q9FNE1 (Q9FNE1) Receptor-like serine/threonine
kinase, partial (33%)
Length = 1110
Score = 65.9 bits (159), Expect = 2e-11
Identities = 57/229 (24%), Positives = 102/229 (43%), Gaps = 7/229 (3%)
Frame = +1
Query: 54 LTSLVNSASFTNYNNFTVPAT-AGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCS 112
+ SL + N+ +V + +GS +YG QC DL++ C C A + ++L
Sbjct: 34 MESLSQLVTSNNWGTHSVKISGSGSSIPIYGFAQCFRDLSHTDCLLCYAASRTRLPRCLP 213
Query: 113 AVRGGALQLEGCFVKYDNVKFL--GVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTS 170
+V + L+GCF++YDN F G + V G + G ++ + ++ V +
Sbjct: 214 SV-SARIYLDGCFLRYDNYSFYSEGTDPSRDAVNCTGVAAGDEAERVELQERVGRVVDNV 390
Query: 171 DGV-YRTWRISAYGDFQGA---AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLA 226
+ R G+ +G AQC L C++CL ++ + +K C G A
Sbjct: 391 VNIAERDGNGFGVGEVEGVYALAQCWNTLGSGGCRECLRKAGREVK-GCLPKKEGRALNA 567
Query: 227 KCYARYSEGGDHSRHSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLS 275
CY RYS + D D+ + + + I+ ++A A+I++ LS
Sbjct: 568 GCYLRYST--QKFYNEDGDAGGGNGFLRRRGVIVAEVLAAAAVIMLALS 708
>BQ452510 similar to GP|6633857|gb|A F2K11.7 {Arabidopsis thaliana}, partial
(10%)
Length = 453
Score = 65.9 bits (159), Expect = 2e-11
Identities = 40/109 (36%), Positives = 58/109 (52%), Gaps = 3/109 (2%)
Frame = +3
Query: 28 FIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTN-YNNFTVPATAGSGDTLYGLYQ 86
F+ C K T S YE + +LL+SL ++A+ T YNN + +T + DT+YGL+
Sbjct: 3 FVSQNCGANKATANSP--YEKNLKTLLSSLSSNATTTLFYNNTVLGSTNTTSDTVYGLFM 176
Query: 87 CRGDLNNDQCYRCVARAVSQLGT--LCSAVRGGALQLEGCFVKYDNVKF 133
CRGD + C CVA A +L + CS + + C V+Y NV F
Sbjct: 177 CRGDXSLRLCKECVANATEKLSSDQSCSLSKQAVMWYAECIVRYSNVGF 323
Score = 40.0 bits (92), Expect = 0.001
Identities = 28/87 (32%), Positives = 42/87 (48%), Gaps = 2/87 (2%)
Frame = +3
Query: 152 LTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSESIQRL 211
L+S+A T L Y T G T + YG F C GD S C++C++ + ++L
Sbjct: 84 LSSNATT----TLFYNNTVLGSTNTTSDTVYGLFM----CRGDXSLRLCKECVANATEKL 239
Query: 212 KTE--CGASTWGEMYLAKCYARYSEGG 236
++ C S M+ A+C RYS G
Sbjct: 240 SSDQSCSLSKQAVMWYAECIVRYSNVG 320
>AW234936
Length = 407
Score = 65.5 bits (158), Expect = 3e-11
Identities = 39/136 (28%), Positives = 64/136 (46%), Gaps = 1/136 (0%)
Frame = +3
Query: 5 LRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKN-TPVSDPVYETGVNSLLTSLVNSASF 63
L LF L +IT ++ ++ CS K TP S Y++ + +LLTSL + A+
Sbjct: 9 LNYLFFLFNFSITKTEAIDNLQ-YLNHSCSSNKTFTPNS--TYQSNLQTLLTSLSSHATT 179
Query: 64 TNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEG 123
+ N T +G+ +YG + CRGD++N C C+ A Q+ C + +
Sbjct: 180 AQFFNTTTGGGDAAGENIYGSFMCRGDVSNHTCQECIKTATQQITVRCLNSKEALIWYHE 359
Query: 124 CFVKYDNVKFLGVEDK 139
C V+Y N F ++
Sbjct: 360 CMVRYSNRCFFSAVEE 407
Score = 37.0 bits (84), Expect = 0.011
Identities = 15/47 (31%), Positives = 25/47 (52%)
Frame = +3
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G+ C GD+S CQ+C+ + Q++ C S ++ +C RYS
Sbjct: 237 GSFMCRGDVSNHTCQECIKTATQQITVRCLNSKEALIWYHECMVRYS 377
>BQ576403 similar to PIR|D85065|D850 receptor protein kinase-like protein
[imported] - Arabidopsis thaliana, partial (3%)
Length = 423
Score = 63.2 bits (152), Expect = 1e-10
Identities = 35/108 (32%), Positives = 53/108 (48%), Gaps = 1/108 (0%)
Frame = +1
Query: 33 CSQPKN-TPVSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQCRGDL 91
CS K TP+S Y + + +LLTSL + A+ + N T +G+T+YG + CRGD+
Sbjct: 13 CSSNKTFTPIS--FYNSNLQTLLTSLSSHATTAQFFNSTTGG-GDTGETIYGSFMCRGDV 183
Query: 92 NNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVEDK 139
N C CV A Q+ C + + C V+Y N F ++
Sbjct: 184 TNHTCQECVKTATQQITLRCPNSKDALIWYHECLVRYSNSCFFSTMEE 327
Score = 35.4 bits (80), Expect = 0.031
Identities = 14/47 (29%), Positives = 25/47 (52%)
Frame = +1
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
G+ C GD++ CQ+C+ + Q++ C S ++ +C RYS
Sbjct: 157 GSFMCRGDVTNHTCQECVKTATQQITLRCPNSKDALIWYHECLVRYS 297
>TC212130 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (30%)
Length = 689
Score = 56.6 bits (135), Expect = 1e-08
Identities = 57/230 (24%), Positives = 96/230 (40%), Gaps = 19/230 (8%)
Frame = +3
Query: 5 LRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFT 64
+ L F+ L P ++S+ F C + ++ Y T +N+LL+SL +S T
Sbjct: 18 IHLFFLCCLFLKFIPQGNASMFHTAF--CDNKEGNYTANSTYNTNLNTLLSSL---SSHT 182
Query: 65 NYNNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEG 123
N F + G D + + CRGD+ +C C+ + + C + L L
Sbjct: 183 EINYFFYNFSYGQNPDKVNAIGLCRGDVEPHECRSCLNDSRVTIKQFCPNQKKALLWLNT 362
Query: 124 --CFVKYDNVKFLGVEDKTMVVKKCGPSIGLTS-DALTRRD----AVLAYLQTSDGV--- 173
C ++Y G+ + PS L + + +T D A+ ++ GV
Sbjct: 363 SKCMLRYSPRSIFGIME-------IEPSQSLMNINNVTEPDKFSQALANLMRNLKGVAAS 521
Query: 174 ------YRTWRISA--YGDFQGAAQCTGDLSPSECQDCLSESIQRLKTEC 215
Y T ++A + G A+CT DLS +C DCL +I ++ T C
Sbjct: 522 GDSRRKYATDNVTASSFQTIYGMAECTPDLSEKDCNDCLDGAISKIPTCC 671
Score = 38.9 bits (89), Expect = 0.003
Identities = 17/44 (38%), Positives = 25/44 (56%)
Frame = +3
Query: 74 TAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGG 117
TA S T+YG+ +C DL+ C C+ A+S++ T C GG
Sbjct: 558 TASSFQTIYGMAECTPDLSEKDCNDCLDGAISKIPTCCQDKIGG 689
>BE608352 similar to PIR|F86177|F861 protein F19P19.1 [imported] -
Arabidopsis thaliana, partial (30%)
Length = 254
Score = 55.5 bits (132), Expect = 3e-08
Identities = 23/63 (36%), Positives = 32/63 (50%)
Frame = +1
Query: 85 YQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVEDKTMVVK 144
+QC+GDL N CY CV+R LC +QL GC++ Y+ F + M+ K
Sbjct: 1 FQCKGDLTNSDCYNCVSRLPVLCDKLCGKTTAARVQLLGCYILYEVAGFSQISGMQMLYK 180
Query: 145 KCG 147
CG
Sbjct: 181 TCG 189
Score = 37.0 bits (84), Expect = 0.011
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = +1
Query: 190 QCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGG 236
QC GDL+ S+C +C+S CG +T + L CY Y G
Sbjct: 4 QCKGDLTNSDCYNCVSRLPVLCDKLCGKTTAARVQLLGCYILYEVAG 144
>CA801618
Length = 446
Score = 55.1 bits (131), Expect = 4e-08
Identities = 37/122 (30%), Positives = 58/122 (47%), Gaps = 1/122 (0%)
Frame = +1
Query: 8 LFILTLSAITTPSTSSSIDTFIFGGCSQPKN-TPVSDPVYETGVNSLLTSLVNSASFTNY 66
LF+L +T TS+S+ + C+ K TP S + T +N+LL+ L S++ TN
Sbjct: 73 LFLLCTLLLTIAETSASVFNNV--SCTSNKTFTPNS--TFNTNLNTLLSYL--SSNVTNN 234
Query: 67 NNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFV 126
F +T+YGLY CRGD+ C CV A + + C + + C +
Sbjct: 235 VRFFNATVGKDSNTVYGLYMCRGDVPFALCRECVGFATQTIPSSCPTSKEAVIWYNECLL 414
Query: 127 KY 128
+Y
Sbjct: 415 RY 420
>AW706364 similar to GP|8777384|dbj| 33 kDa secretory protein-like
{Arabidopsis thaliana}, partial (30%)
Length = 396
Score = 51.6 bits (122), Expect = 4e-07
Identities = 31/103 (30%), Positives = 45/103 (43%)
Frame = +3
Query: 80 TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVEDK 139
TLY L QC DL+ C +C++ AV+ CS +G + C+V+Y+ F D
Sbjct: 57 TLYALVQCTRDLSEISCAQCLSIAVNNFPNFCSNRKGCRVLYSSCYVRYELYPFFFPLDS 236
Query: 140 TMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAY 182
K GPS T A+L Y+ + W +S Y
Sbjct: 237 ----NKTGPSNTAKVSVYT*LKAMLPYMPKMAFLLNAWMVSNY 353
Score = 35.4 bits (80), Expect = 0.031
Identities = 16/54 (29%), Positives = 22/54 (40%)
Frame = +3
Query: 179 ISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
+S + QCT DLS C CLS ++ C + + CY RY
Sbjct: 42 LSPFVTLYALVQCTRDLSEISCAQCLSIAVNNFPNFCSNRKGCRVLYSSCYVRY 203
>TC224215 weakly similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase
homolog RK20-1, partial (17%)
Length = 489
Score = 50.8 bits (120), Expect = 7e-07
Identities = 25/80 (31%), Positives = 41/80 (51%)
Frame = +3
Query: 73 ATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVK 132
ATA + +YGL QC DL+ +C C+ A+S++ C+ GG + C ++Y+N
Sbjct: 162 ATATNIQAIYGLVQCTPDLSQPECKHCLIGAISEIPRCCNGKIGGRVLRPSCNIRYENYP 341
Query: 133 FLGVEDKTMVVKKCGPSIGL 152
F ++ T PS+ L
Sbjct: 342 F--YDEPTAYAPAPSPSLSL 395
Score = 37.4 bits (85), Expect = 0.008
Identities = 17/46 (36%), Positives = 21/46 (44%)
Frame = +3
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
G QCT DLS EC+ CL +I + C G + C RY
Sbjct: 192 GLVQCTPDLSQPECKHCLIGAISEIPRCCNGKIGGRVLRPSCNIRY 329
>BU762295 weakly similar to PIR|D85065|D850 receptor protein kinase-like
protein [imported] - Arabidopsis thaliana, partial (10%)
Length = 426
Score = 48.9 bits (115), Expect = 3e-06
Identities = 21/68 (30%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Frame = +2
Query: 67 NNFTVPATAGSG-DTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
+ F +G G +T+YGL QC DL+ +C C+ A+S++ C + +GG + C
Sbjct: 20 HKFAAANVSGPGFETIYGLVQCTPDLSEQECTSCLVDAISEIPRCCDSKKGGRVVRPSCN 199
Query: 126 VKYDNVKF 133
+Y+ F
Sbjct: 200 FRYETYPF 223
Score = 38.5 bits (88), Expect = 0.004
Identities = 17/46 (36%), Positives = 22/46 (46%)
Frame = +2
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
G QCT DLS EC CL ++I + C + G + C RY
Sbjct: 71 GLVQCTPDLSEQECTSCLVDAISEIPRCCDSKKGGRVVRPSCNFRY 208
>BI424423 weakly similar to GP|4530126|gb|A receptor-like protein kinase
homolog RK20-1 {Phaseolus vulgaris}, partial (9%)
Length = 429
Score = 48.1 bits (113), Expect = 5e-06
Identities = 20/61 (32%), Positives = 32/61 (51%)
Frame = +1
Query: 73 ATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVK 132
ATA + T+YG QC DL+ C+ C+ A S++ + C G + C + Y+N +
Sbjct: 214 ATAPNSQTIYGAAQCTPDLSEQDCHSCLVEAFSRITSCCIGNISGRVAAPSCNITYENFR 393
Query: 133 F 133
F
Sbjct: 394 F 396
Score = 41.6 bits (96), Expect = 4e-04
Identities = 21/62 (33%), Positives = 30/62 (47%), Gaps = 2/62 (3%)
Frame = +1
Query: 187 GAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGG--DHSRHSDD 244
GAAQCT DLS +C CL E+ R+ + C + G + C Y D H+ +
Sbjct: 244 GAAQCTPDLSEQDCHSCLVEAFSRITSCCIGNISGRVAAPSCNITYENFRFYDEHEHATE 423
Query: 245 DS 246
D+
Sbjct: 424 DA 429
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,981,339
Number of Sequences: 63676
Number of extensions: 196985
Number of successful extensions: 1105
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 1031
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1080
length of query: 292
length of database: 12,639,632
effective HSP length: 96
effective length of query: 196
effective length of database: 6,526,736
effective search space: 1279240256
effective search space used: 1279240256
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0097b.12


