

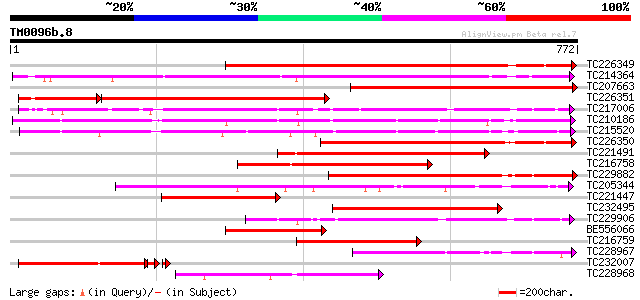
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.8
(772 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 625 e-179
TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete 599 e-171
TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 512 e-145
TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 411 e-144
TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete 508 e-144
TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete 486 e-137
TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type proteas... 468 e-132
TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 452 e-127
TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 452 e-127
TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase... 327 2e-89
TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin... 307 1e-83
TC205344 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (... 280 2e-75
TC221447 similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like p... 268 5e-72
TC232495 similar to UP|Q8W554 (Q8W554) AT3g14240/MLN21_2, partia... 263 2e-70
TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like pr... 261 8e-70
BE556066 238 1e-62
TC216759 similar to PIR|T05768|T05768 subtilisin-like proteinase... 231 9e-61
TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 214 1e-55
TC232007 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 202 4e-55
TC228968 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 211 7e-55
>TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(56%)
Length = 1773
Score = 625 bits (1612), Expect = e-179
Identities = 307/480 (63%), Positives = 376/480 (77%), Gaps = 2/480 (0%)
Frame = +1
Query: 294 DYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITL 353
DY+RD +AIGAF+A GILVS SAGN GP P SLSN APWITTVGAGT+DRDFPAY+ L
Sbjct: 1 DYYRDSVAIGAFSAMEKGILVSCSAGNSGPGPYSLSNVAPWITTVGAGTLDRDFPAYVAL 180
Query: 354 GNNITHTGASLYRGKPLSDSPLPLVYAGNASNFSV-GYLCLPDSLVPSKVLGKIVICERG 412
GN + +G SLYRG L DS LPLVYAGN SN ++ G LC+ +L P KV GKIV+C+RG
Sbjct: 181 GNGLNFSGVSLYRGNALPDSSLPLVYAGNVSNGAMNGNLCITGTLSPEKVAGKIVLCDRG 360
Query: 413 GNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRN 472
ARV+KG VVK AG +GM+L+N GEELVAD+HLLPA A+G+++ A+K Y+ S
Sbjct: 361 LTARVQKGSVVKSAGALGMVLSNTAANGEELVADAHLLPATAVGQKAGDAIKKYLVSDAK 540
Query: 473 PTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGL 532
PT K+ F GT + ++PSPVVAAFSSRGPN +TP+ILKPDLIAPGVNILAGW+ A+GPTGL
Sbjct: 541 PTVKIFFEGTKVGIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGL 720
Query: 533 PVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQ 592
PVD R V FNIISGTSMSCPHVSGLAA++K +HP+WSPAA+RSALMTT+YT YK G+ +Q
Sbjct: 721 PVDNRRVDFNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYTVYKTGEKLQ 900
Query: 593 DVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFK 652
D ATGKP+TP D G+GHVDPVA+L+PGLVYD VDDYLGFLCALNY++ EI ++R F+
Sbjct: 901 DSATGKPSTPFDHGSGHVDPVAALNPGLVYDLTVDDYLGFLCALNYSAAEISTLAKRKFQ 1080
Query: 653 CDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQS 712
CD K+Y V D NYPSFAV E++ + VK++RTLTNVG GTYKASV+S +
Sbjct: 1081CDAGKQYSVTDLNYPSFAVLFESSGSV---------VKHTRTLTNVGPAGTYKASVTSDT 1233
Query: 713 PSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTK-SFAYLYWSDGKHRVASPIAITW 771
SVKI+VEPQ+L F+E EKK++TVTF+S+ P T+ +F + WSDGKH V SPI++ W
Sbjct: 1234ASVKISVEPQVLSFKE-NEKKTFTVTFSSSGSPQHTENAFGRVEWSDGKHLVGSPISVNW 1410
>TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete
Length = 2667
Score = 599 bits (1544), Expect = e-171
Identities = 339/790 (42%), Positives = 481/790 (59%), Gaps = 24/790 (3%)
Frame = +1
Query: 4 PIFQLLQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKST-----MPATFND----H 54
P L LL+ L+ S+ + KK+Y++++ + FN H
Sbjct: 52 PSIHFLLQILLVCLLHRPSFAV---------KKSYVVYLGAHSHGPELSSVDFNQVTQSH 204
Query: 55 QHWFDSSLQSVSESAE-ILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHT 113
+ S L S + + + I Y+Y +GF+ L + A +A+ P VLSV +LHT
Sbjct: 205 HDFLGSFLGSSNTAKDSIFYSYTRHINGFAATLDEEVAVEIAKHPKVLSVFENRGRKLHT 384
Query: 114 TRTPEFLGLLKKTTTLSPGSDKQSQ----VVIGVLDTGVWPELKSLDDTGLSPVPSTWKG 169
TR+ +F+ L S K+++ V+IG LDTGVWPE KS + GL P+PS W+G
Sbjct: 385 TRSWDFMELEHNGVIQSSSIWKKARFGEGVIIGNLDTGVWPESKSFSEQGLGPIPSKWRG 564
Query: 170 QCEAGNNMNSSSCNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGS 229
C G + ++ CNRKLIGAR+F+KGY + GP++ S +S RD++GHG+HTL+TA G+
Sbjct: 565 ICHNGID-HTFHCNRKLIGARYFNKGYASVAGPLNSSFDS--PRDNEGHGTHTLSTAGGN 735
Query: 230 AVAGASLFGLASGTARGMATQARVAAYKVCW--LGG--CFSSDIAAGIDKAIEDGVNIIS 285
VA S+FG GTA+G + ARVAAYKVCW + G CF +DI A D AI DGV+++S
Sbjct: 736 MVARVSVFGQGHGTAKGGSPMARVAAYKVCWPPVAGDECFDADILAAFDLAIHDGVDVLS 915
Query: 286 MSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDR 345
+S+GGSS+ +F+D +AIG+F A G++V SAGN GP+ ++ N APW TV A T+DR
Sbjct: 916 VSLGGSSSTFFKDSVAIGSFHAAKRGVVVVCSAGNSGPAEATAENLAPWHVTVAASTMDR 1095
Query: 346 DFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNFSV----GYLCLPDSLVPSK 401
FP Y+ LGN+IT G SL K L+ P++ A +A S LC +L P+K
Sbjct: 1096QFPTYVVLGNDITFKGESLSATK-LAHKFYPIIKATDAKLASARAEDAVLCQNGTLDPNK 1272
Query: 402 VLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSK 461
GKIV+C RG NARV+KG AG +GM+LAN++ G E++AD H+LPA+ +
Sbjct: 1273AKGKIVVCLRGINARVDKGEQAFLAGAVGMVLANDKTTGNEIIADPHVLPASHINFTDGS 1452
Query: 462 ALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILA 521
A+ Y+ S++ P A + T L KP+P +AAFSS+GPN + P+ILKPD+ APGV+++A
Sbjct: 1453AVFKYINSTKFPVAYITHPKTQLDTKPAPFMAAFSSKGPNTIVPEILKPDITAPGVSVIA 1632
Query: 522 GWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTS 581
+T A GPT D R + FN +SGTSMSCPHVSG+ +L+ +P WS AAI+SA+MTT+
Sbjct: 1633AYTEAQGPTNQVFDKRRIPFNSVSGTSMSCPHVSGIVGLLRALYPTWSTAAIKSAIMTTA 1812
Query: 582 YTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSL 641
T + + + GK ATP +GAGHV P ++DPGLVYD +DDYL FLCAL Y
Sbjct: 1813TTLDNEVEPLLNATDGK-ATPFSYGAGHVQPNRAMDPGLVYDITIDDYLNFLCALGYNET 1989
Query: 642 EIKLASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTP 701
+I + + +KC +KK+ + + NYPS VP + S V +RTL NVG+P
Sbjct: 1990QISVFTEGPYKC--RKKFSLLNLNYPSITVPKLSGS-----------VTVTRTLKNVGSP 2130
Query: 702 GTYKASVSSQSP-SVKIAVEPQILRFQELYEKKSYTVTFTS-NSMPSGTKSFAYLYWSDG 759
GTY A V Q+P + ++V+P IL+F+ + E+KS+ +TF + + +F L WSDG
Sbjct: 2131GTYIAHV--QNPYGITVSVKPSILKFKNVGEEKSFKLTFKAMQGKATNNYAFGKLIWSDG 2304
Query: 760 KHRVASPIAI 769
KH V SPI +
Sbjct: 2305KHYVTSPIVV 2334
>TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (37%)
Length = 1171
Score = 512 bits (1318), Expect = e-145
Identities = 248/310 (80%), Positives = 273/310 (88%), Gaps = 1/310 (0%)
Frame = +1
Query: 464 KDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGW 523
++YV SS NPTAK+ F GTHLQV+PSPVVAAFSSRGPN LTPKILKPDLIAPGVNILAGW
Sbjct: 4 RNYVSSSPNPTAKIAFLGTHLQVQPSPVVAAFSSRGPNALTPKILKPDLIAPGVNILAGW 183
Query: 524 TGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYT 583
TGA+GPTGL VDTRHVSFNIISGTSMSCPHVSGLAAILKG+HP+WSPAAIRSALMTT+YT
Sbjct: 184 TGAVGPTGLTVDTRHVSFNIISGTSMSCPHVSGLAAILKGAHPQWSPAAIRSALMTTAYT 363
Query: 584 AYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEI 643
+YKNG+TIQD++TG+P TP D+GAGHVDPVA+LDPGLVYDANVDDYLGF CALNY+S +I
Sbjct: 364 SYKNGETIQDISTGQPGTPFDYGAGHVDPVAALDPGLVYDANVDDYLGFFCALNYSSFQI 543
Query: 644 KLASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGT 703
KLA+RRD+ CDPKK YRVEDFNYPSFAVP++TASGIGGGS TVKYSR LTNVG PGT
Sbjct: 544 KLAARRDYTCDPKKDYRVEDFNYPSFAVPMDTASGIGGGSDTLKTVKYSRVLTNVGAPGT 723
Query: 704 YKASVSSQSPS-VKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLYWSDGKHR 762
YKASV S S VK VEP L F ELYEKK YTV+FT SMPSGT SFA L W+DGKH+
Sbjct: 724 YKASVMSLGDSNVKTVVEPNTLSFTELYEKKDYTVSFTYTSMPSGTTSFARLEWTDGKHK 903
Query: 763 VASPIAITWT 772
V SPIA +WT
Sbjct: 904 VGSPIAFSWT 933
>TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(48%)
Length = 1490
Score = 411 bits (1056), Expect(2) = e-144
Identities = 204/313 (65%), Positives = 246/313 (78%), Gaps = 2/313 (0%)
Frame = +3
Query: 125 KTTTLSPGSDKQSQVVIGVLDTG-VWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCN 183
+ T SP S V+IGVL+T + + + +TGL PVPSTWKG CE G N+ +S+CN
Sbjct: 552 RAPTCSPSRGSGSDVIIGVLNTWRLAGKAGASTNTGLGPVPSTWKGACETGTNLTASNCN 731
Query: 184 RKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGT 243
RKLIGARFFSKG EA LGPI+ + ESRSARDDDGHG+HT +TAAGS V+ ASLFG ASGT
Sbjct: 732 RKLIGARFFSKGVEAILGPINETEESRSARDDDGHGTHTASTAAGSVVSDASLFGYASGT 911
Query: 244 ARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIG 303
ARGMAT+ARVAAYKVCW GGCFSSDI A I++AI D VN++S+S+GG +DY+RD +AIG
Sbjct: 912 ARGMATRARVAAYKVCWKGGCFSSDILAAIERAILDNVNVLSLSLGGGMSDYYRDSVAIG 1091
Query: 304 AFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGAS 363
AF+A +GILVS SAGN GPSP SLSN APWITTVGAGT+DRDFPAY+ LGN + +G S
Sbjct: 1092AFSAMENGILVSCSAGNAGPSPYSLSNVAPWITTVGAGTLDRDFPAYVALGNGLNFSGVS 1271
Query: 364 LYRGKPLSDSPLPLVYAGNASNFSV-GYLCLPDSLVPSKVLGKIVICERGGNARVEKGLV 422
LYRG + DSPLP VYAGN SN ++ G LC+ +L P KV GKIV+C+RG ARV+KG V
Sbjct: 1272LYRGNAVPDSPLPFVYAGNVSNGAMNGNLCITGTLSPEKVAGKIVLCDRGLTARVQKGSV 1451
Query: 423 VKRAGGIGMILAN 435
VK AG +GM+L+N
Sbjct: 1452VKSAGALGMVLSN 1490
Score = 120 bits (302), Expect(2) = e-144
Identities = 59/113 (52%), Positives = 80/113 (70%)
Frame = +2
Query: 12 ALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQSVSESAEI 71
A+L +++F + AE + K TYI+H+ KS MP +F H W++SSL++VS+SAEI
Sbjct: 230 AILWVVLFLGLHEAAEPE-----KSTYIVHVAKSEMPESFEHHALWYESSLKTVSDSAEI 394
Query: 72 LYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLK 124
+YTY + HG++TRLT +EA L Q G+L+V PE RYELHTTRTP FLGL K
Sbjct: 395 MYTYDNAIHGYATRLTAEEARVLETQAGILAVLPETRYELHTTRTPMFLGLDK 553
>TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete
Length = 2422
Score = 508 bits (1307), Expect = e-144
Identities = 322/780 (41%), Positives = 438/780 (55%), Gaps = 23/780 (2%)
Frame = +3
Query: 13 LLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQH---WFDSSLQSVSES- 68
LL+LL F S I +A Q K +YI++ S ND + S LQ V++S
Sbjct: 54 LLMLLCFASFLQICH--SASQLK-SYIVYTGNS-----MNDEASALTLYSSMLQEVADSN 209
Query: 69 AE---ILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKK 125
AE + + +K GF LT +EA+ +A V++V P + +LHTTR+ +F+G +
Sbjct: 210 AEPKLVQHHFKRSFSGFVAMLTEEEADRMARHDRVVAVFPNKKKQLHTTRSWDFIGFPLQ 389
Query: 126 TTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRK 185
SD V+I V D+G+WPE +S +D G P PS WKG C+ N +CN K
Sbjct: 390 ANRAPAESD----VIIAVFDSGIWPESESFNDKGFGPPPSKWKGTCQTSKNF---TCNNK 548
Query: 186 LIGAR------FFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGL 239
+IGA+ FFSK + +S RD DGHG+H +TAAG+ V+ AS+ GL
Sbjct: 549 IIGAKIYKVDGFFSKD------------DPKSVRDIDGHGTHVASTAAGNPVSTASMLGL 692
Query: 240 ASGTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGG-SSADYFRD 298
GT+RG T+AR+A YKVCW GC +DI A D AI DGV+II++S+GG S +YFRD
Sbjct: 693 GQGTSRGGVTKARIAVYKVCWFDGCTDADILAAFDDAIADGVDIITVSLGGFSDENYFRD 872
Query: 299 IIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNIT 358
IAIGAF A +G+L TSAGN GP PSSLSN +PW +V A TIDR F + LGN IT
Sbjct: 873 GIAIGAFHAVRNGVLTVTSAGNSGPRPSSLSNFSPWSISVAASTIDRKFVTKVELGNKIT 1052
Query: 359 HTGASLYRGKPLSDSPLPLVYAGNASNFSVGY------LCLPDSLVPSKVLGKIVICERG 412
+ G S+ L P++Y G+A N G C SL V GKIV+CE
Sbjct: 1053YEGTSI-NTFDLKGELYPIIYGGDAPNKGEGIDGSSSRYCSSGSLDKKLVKGKIVLCESR 1229
Query: 413 GNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRN 472
K L AG +G ++ G + S LP + L + ++ DY+ S+R
Sbjct: 1230S-----KALGPFDAGAVGALIQGQ---GFRDLPPSLPLPGSYLALQDGASVYDYINSTRT 1385
Query: 473 PTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGL 532
P A +F + +PVVA+FSSRGPN +TP+ILKPDL+APGV+ILA W+ A P+ +
Sbjct: 1386PIAT-IFKTDETKDTIAPVVASFSSRGPNIVTPEILKPDLVAPGVSILASWSPASPPSDV 1562
Query: 533 PVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQ 592
D R ++FNIISGTSM+CPHVSG AA +K HP WSPAAIRSALMTT+ +
Sbjct: 1563EGDNRTLNFNIISGTSMACPHVSGAAAYVKSFHPTWSPAAIRSALMTTAKQLSPKTHLLA 1742
Query: 593 DVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFK 652
+ A +GAG +DP ++ PGLVYDA DY+ FLC Y++ ++L + +
Sbjct: 1743EFA---------YGAGQIDPSKAVYPGLVYDAGEIDYVRFLCGQGYSTRTLQLITGDNSS 1895
Query: 653 CDPKKKYRVEDFNYPSFA--VPLETASGIGGGSHAPITVKYSRTLTNVGTP-GTYKASVS 709
C K D NY SFA VP ++ + G ++RT+TNVG+P TYKA+V+
Sbjct: 1896CPETKNGSARDLNYASFALFVPPYNSNSVSG--------SFNRTVTNVGSPKSTYKATVT 2051
Query: 710 SQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLYWSDGKHRVASPIAI 769
S +KI V P +L F L +K+++ +T T G L W DGK++V SPI +
Sbjct: 2052SPK-GLKIEVNPSVLPFTSLNQKQTFVLTITGKL--EGPIVSGSLVWDDGKYQVRSPIVV 2222
>TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete
Length = 2401
Score = 486 bits (1252), Expect = e-137
Identities = 310/785 (39%), Positives = 443/785 (55%), Gaps = 19/785 (2%)
Frame = +1
Query: 5 IFQLLQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDK--STMPATFNDHQHWFDSSL 62
+ L + LL L + SS K+ YI++M ST + NDH +S L
Sbjct: 82 LLHLFYTLLLFLGVRSSSSAGNGSNDVTNRKEVYIVYMGAADSTDASFRNDHAQVLNSVL 261
Query: 63 QSVSESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGL 122
+ +E+A ++ YKH GF+ RL+ +EA ++A++PGV+SV P +LHTTR+ +FL
Sbjct: 262 RR-NENA-LVRNYKHGFSGFAARLSKKEATSIAQKPGVVSVFPGPVLKLHTTRSWDFLKY 435
Query: 123 LKKTTT-LSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSS 181
+ P + +S VIG+LDTG+WPE S D G+ PVPS WKG C + SS+
Sbjct: 436 QTQVKIDTKPNAVSKSSSVIGILDTGIWPEAASFSDKGMGPVPSRWKGTCMKSQDFYSSN 615
Query: 182 CNRKLIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLAS 241
CNRKLIGAR+++ P D + +ARD +GHG+H TAAG V AS +G+A+
Sbjct: 616 CNRKLIGARYYAD-------PND--SGDNTARDSNGHGTHVAGTAAGVMVTNASYYGVAT 768
Query: 242 GTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSA---DYFRD 298
G A+G + ++R+A Y+VC GC S I A D AI DGV+++S+S+G S+ D D
Sbjct: 769 GCAKGGSPESRLAVYRVCSNFGCRGSSILAAFDDAIADGVDLLSVSLGASTGFRPDLTSD 948
Query: 299 IIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNIT 358
I++GAF A HGILV SAGN GPS +L N APWI TV A TIDR+F + I LG+N
Sbjct: 949 PISLGAFHAMEHGILVVCSAGNDGPSSYTLVNDAPWILTVAASTIDRNFLSNIVLGDNKI 1128
Query: 359 HTGASLYRGKPLSDSP-LPLVYAGNASNFSVGYL----CLPDSLVPSKVLGKIVICERGG 413
G ++ PLS+SP PL+Y +A S + C P+SL +KV GKIV+C+
Sbjct: 1129IKGKAI-NLSPLSNSPKYPLIYGESAKANSTSLVEARQCHPNSLDGNKVKGKIVVCDDKN 1305
Query: 414 N--ARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSR 471
+ + +K VK GGIG++ ++ E + ++ PA + + + Y+ S+
Sbjct: 1306DKYSTRKKVATVKAVGGIGLVHITDQ--NEAIASNYGDFPATVISSKDGVTILQYINSTS 1479
Query: 472 NPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTG 531
NP A ++ + L KP+P+V FSSRGP+ L+ ILKPD+ APGVNILA W G G
Sbjct: 1480NPVATILATTSVLDYKPAPLVPNFSSRGPSSLSSNILKPDIAAPGVNILAAWIGN-GTEV 1656
Query: 532 LPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTI 591
+P + + IISGTSM+CPHVSGLA+ +K +P WS ++I+SA+MT++ + N +
Sbjct: 1657VPKGKKPSLYKIISGTSMACPHVSGLASSVKTRNPTWSASSIKSAIMTSAIQS-NNLKAP 1833
Query: 592 QDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASR--- 648
+G ATP D+GAG + L PGLVY+ + DYL FLC + + +K+ S+
Sbjct: 1834ITTESGSVATPYDYGAGEMTTSEPLQPGLVYETSSVDYLNFLCYIGFNVTTVKVISKTVP 2013
Query: 649 RDFKCDPK--KKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKA 706
R+F C PK + + NYPS A+ G A V SRT+TNVG
Sbjct: 2014RNFNC-PKDLSSDHISNINYPSIAINF-------SGKRA---VNLSRTVTNVGEDDETVY 2160
Query: 707 SVSSQSPS-VKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLYWSDGKHRVAS 765
S +PS V + + P LRF + +K SY V F+S F + WS+GK+ V S
Sbjct: 2161SPIVDAPSGVHVTLTPNKLRFTKSSKKLSYRVIFSSTLTSLKEDLFGSITWSNGKYMVRS 2340
Query: 766 PIAIT 770
P +T
Sbjct: 2341PFVLT 2355
>TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type protease precursor
{Glycine max;} , complete
Length = 2392
Score = 468 bits (1205), Expect = e-132
Identities = 310/787 (39%), Positives = 437/787 (55%), Gaps = 30/787 (3%)
Frame = +2
Query: 14 LLLLIFCSSYTIAEKKTAQQAKKTYIIHMDK--STMPATFNDHQHWFDSSLQSVSESAEI 71
L L++ SS T E +K+ YI++M ST + N+H +S L+ +E+A +
Sbjct: 77 LFLVVSSSSSTGNESNDDTNSKEVYIVYMGAADSTKASLKNEHAQILNSVLRR-NENA-L 250
Query: 72 LYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFL------GLLKK 125
+ YKH GF+ RL+ +EA ++A++PGV+SV P+ +LHTTR+ +FL + K
Sbjct: 251 VRNYKHGFSGFAARLSKEEANSIAQKPGVVSVFPDPILKLHTTRSWDFLKSQTRVNIDTK 430
Query: 126 TTTLSPGSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRK 185
TLS S S V++GVLDTG+WPE S D G PVPS WKG C + NSS CNRK
Sbjct: 431 PNTLSGSSFSSSDVILGVLDTGIWPEAASFSDKGFGPVPSRWKGTCMTSKDFNSSCCNRK 610
Query: 186 LIGARFFSKGYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTAR 245
+IGARF+ + E ++ARD +GHG+H +TA G V+GAS +GLA+GTAR
Sbjct: 611 IIGARFYP------------NPEEKTARDFNGHGTHVSSTAVGVPVSGASFYGLAAGTAR 754
Query: 246 GMATQARVAAYKVCW-LGGCFSSDIAAGIDKAIEDGVNIISMSI---GGSSADYFRDIIA 301
G + ++R+A YKVC G C S I AG D AI DGV+I+S+S+ GG+ D D IA
Sbjct: 755 GGSPESRLAVYKVCGAFGSCPGSAILAGFDDAIHDGVDILSLSLGGFGGTKTDLTTDPIA 934
Query: 302 IGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTG 361
IGAF + GILV +AGN G P ++ N APWI TV A TIDRD + + LGNN G
Sbjct: 935 IGAFHSVQRGILVVCAAGNDG-EPFTVLNDAPWILTVAASTIDRDLQSDVVLGNNQVVKG 1111
Query: 362 ASLYRGKPLSDSPLPLVYA-----GNASNFSVGYLCLPDSLVPSKVLGKIVICERGGN-- 414
++ L+ P++YA N SN + C PDSL P KV+GKIV+C+ G N
Sbjct: 1112RAINFSPLLNSPDYPMIYAESAARANISNITDARQCHPDSLDPKKVIGKIVVCD-GKNDI 1288
Query: 415 --ARVEKGLVVKRAGGIGMILANNEEFGEELVADSHL-LPAAALGERSSKALKDYVFSSR 471
+ EK ++VK GGIG++ ++ VA ++ P + + A+ Y+ S+
Sbjct: 1289YYSTDEKIVIVKALGGIGLVHITDQSGS---VAFYYVDFPVTEVKSKHGDAILQYINSTS 1459
Query: 472 NPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTG 531
+P ++ T KP+P V FSSRGP+ +T +LKPD+ APGVNILA W G +
Sbjct: 1460HPVGTILATVTIPDYKPAPRVGYFSSRGPSLITSNVLKPDIAAPGVNILAAWFGN-DTSE 1636
Query: 532 LPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTI 591
+P + + I+SGTSM+ PHVSGLA +K +P WS +AI+SA+MT +A +N
Sbjct: 1637VPKGRKPSLYRILSGTSMATPHVSGLACSVKRKNPTWSASAIKSAIMT---SAIQNDNLK 1807
Query: 592 QDVAT--GKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLC--ALNYTSLEIKLAS 647
+ T G ATP D+GAG + L PGLVY+ N DYL +LC LN T +++ +
Sbjct: 1808GPITTDSGLIATPYDYGAGAITTSEPLQPGLVYETNNVDYLNYLCYNGLNITMIKVISGT 1987
Query: 648 -RRDFKCDPKKKYR--VEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTY 704
+F C PK + NYPS AV G + A + SRT+TNV
Sbjct: 1988VPENFNC-PKDSSSDLISSINYPSIAV------NFTGKADAVV----SRTVTNVDEEDET 2134
Query: 705 KASVSSQSPS-VKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLYWSDGKHRV 763
++PS V + + P L F +K+SY +TF + F + WS+ K+ V
Sbjct: 2135VYFPVVEAPSEVIVTLFPYNLEFTTSIKKQSYNITFRPKTSLK-KDLFGSITWSNDKYMV 2311
Query: 764 ASPIAIT 770
P +T
Sbjct: 2312RIPFVLT 2332
>TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 1312
Score = 452 bits (1164), Expect = e-127
Identities = 223/349 (63%), Positives = 274/349 (77%), Gaps = 1/349 (0%)
Frame = +1
Query: 424 KRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTH 483
+ AG +GM+L+N GEELVAD+HLLPA A+G+++ A+K Y+FS PT K++F GT
Sbjct: 4 RSAGALGMVLSNTAANGEELVADAHLLPATAVGQKAGDAIKKYLFSDAKPTVKILFEGTK 183
Query: 484 LQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNI 543
L ++PSPVVAAFSSRGPN +TP+ILKPDLIAPGVNILAGW+ A+GPTGLPVD R V FNI
Sbjct: 184 LGIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLPVDNRRVDFNI 363
Query: 544 ISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPL 603
ISGTSMSCPHVSGLAA++K +HP+WSPAA+RSALMTT+YT YK G+ +QD ATGKP+TP
Sbjct: 364 ISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYTVYKTGEKLQDSATGKPSTPF 543
Query: 604 DFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKYRVED 663
D G+GHVDPVA+L+PGLVYD VDDYLGFLCALNY++ EI ++R F+CD K+Y V D
Sbjct: 544 DHGSGHVDPVAALNPGLVYDLTVDDYLGFLCALNYSASEINTLAKRKFQCDAGKQYSVTD 723
Query: 664 FNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAVEPQI 723
NYPSFAV E+ GG VK++RTLTNVG GTYKASV+S SVKI+VEPQ+
Sbjct: 724 LNYPSFAVLFES----GG------VVKHTRTLTNVGPAGTYKASVTSDMASVKISVEPQV 873
Query: 724 LRFQELYEKKSYTVTFTSNSMP-SGTKSFAYLYWSDGKHRVASPIAITW 771
L F+E EKKS+TVTF+S+ P +F + WSDGKH V +PI+I W
Sbjct: 874 LSFKE-NEKKSFTVTFSSSGSPQQRVNAFGRVEWSDGKHVVGTPISINW 1017
>TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (34%)
Length = 872
Score = 452 bits (1163), Expect = e-127
Identities = 224/289 (77%), Positives = 248/289 (85%)
Frame = +3
Query: 365 YRGKPLSDSPLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVK 424
Y GK +SPLP+VYA N S+ S LC +L+ KV GKIVIC+RGGNARVEKGLVVK
Sbjct: 3 YNGKLPPNSPLPIVYAANVSDESQN-LCTRGTLIAEKVAGKIVICDRGGNARVEKGLVVK 179
Query: 425 RAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHL 484
AGGIGMIL+NNE++GEELVADS+LLPAAALG++SS LK YVFSS NPTAKL FGGT L
Sbjct: 180 SAGGIGMILSNNEDYGEELVADSYLLPAAALGQKSSNELKKYVFSSPNPTAKLGFGGTQL 359
Query: 485 QVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNII 544
V+PSPVVAAFSSRGPN LTPKILKPDLIAPGVNILAGWTGA+GPTGL DTRHV FNII
Sbjct: 360 GVQPSPVVAAFSSRGPNVLTPKILKPDLIAPGVNILAGWTGAVGPTGLTEDTRHVEFNII 539
Query: 545 SGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLD 604
SGTSMSCPHV+GLAA+LKG+HPEWSPAAIRSALMTT+Y YKNGQTI+DVATG PATP D
Sbjct: 540 SGTSMSCPHVTGLAALLKGTHPEWSPAAIRSALMTTAYRTYKNGQTIKDVATGLPATPFD 719
Query: 605 FGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKC 653
+GAGHVDPVA+ DPGLVYD VDDYL F CALNY+ +IKL +RRDF C
Sbjct: 720 YGAGHVDPVAAFDPGLVYDTTVDDYLSFFCALNYSPYQIKLVARRDFTC 866
>TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(35%)
Length = 796
Score = 327 bits (837), Expect = 2e-89
Identities = 163/265 (61%), Positives = 196/265 (73%)
Frame = +3
Query: 311 GILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPL 370
G+ VS+SAGN GPS S++N APW+TTVGAGTIDRDFP+ + LG+ +G SLY G L
Sbjct: 3 GVFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQVILGDGRRLSGVSLYAGAAL 182
Query: 371 SDSPLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIG 430
LVY G S LC+ +SL P+ V GKIVIC+RG + RV KGLVVK+AGG+G
Sbjct: 183 KGKMYQLVYPGK-SGILGDSLCMENSLDPNMVKGKIVICDRGSSPRVAKGLVVKKAGGVG 359
Query: 431 MILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSP 490
MILAN GE LV D+HLLPA A+G +K Y+ SS NPTA L F GT L +KP+P
Sbjct: 360 MILANGISNGEGLVGDAHLLPACAVGANEGDVIKKYISSSTNPTATLDFKGTILGIKPAP 539
Query: 491 VVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMS 550
V+A+FS+RGPNGL P+ILKPD IAPGVNILA WT A+GPTGL DTR FNI+SGTSM+
Sbjct: 540 VIASFSARGPNGLNPQILKPDFIAPGVNILAAWTQAVGPTGLDSDTRRTEFNILSGTSMA 719
Query: 551 CPHVSGLAAILKGSHPEWSPAAIRS 575
CPHVSG AA+LK +HP+WSPAA+RS
Sbjct: 720 CPHVSGAAALLKSAHPDWSPAALRS 794
>TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like
proteinase, partial (27%)
Length = 1263
Score = 307 bits (786), Expect = 1e-83
Identities = 171/342 (50%), Positives = 220/342 (64%), Gaps = 3/342 (0%)
Frame = +1
Query: 434 ANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVA 493
AN GEELVADSHLLPA A+G ++ Y S NPT L F GT L VKPSPVVA
Sbjct: 1 ANTAASGEELVADSHLLPAVAVGRIVGDQIRAYASSDPNPTVHLDFRGTVLNVKPSPVVA 180
Query: 494 AFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPH 553
AFSSRGPN +T +ILKPD+I PGVNILAGW+ AIGP+GL DTR FNI+SGTSMSCPH
Sbjct: 181 AFSSRGPNMVTRQILKPDVIGPGVNILAGWSEAIGPSGLSDDTRKTQFNIMSGTSMSCPH 360
Query: 554 VSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPV 613
+SGLAA+LK +HP+WS +AI+SALMTT+ ++D A G + P GAGHV+P
Sbjct: 361 ISGLAALLKAAHPQWSSSAIKSALMTTADVHDNTKSQLRDAAGGAFSNPWAHGAGHVNPH 540
Query: 614 ASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKYRVEDFNYPSFAVPL 673
+L PGLVYDA DY+ FLC+L YT I+L ++R K+ NYPSF+V
Sbjct: 541 KALSPGLVYDATPSDYIKFLCSLEYTPERIQLITKRSGVNCTKRFSDPGQLNYPSFSV-- 714
Query: 674 ETASGIGGGSHAPITVKYSRTLTNVGTPGT-YKASVSSQSPSVKIAVEPQILRFQELYEK 732
+ GG V+Y+R LTNVG G+ Y +V + S +V + V+P L F ++ E+
Sbjct: 715 -----LFGGKR---VVRYTRVLTNVGEAGSVYNVTVDAPS-TVTVTVKPAALVFGKVGER 867
Query: 733 KSYTVTFTS-NSMPSGTK-SFAYLYWSDGKHRVASPIAITWT 772
+ YT TF S N + + F + WS+ +H+V SP+A +WT
Sbjct: 868 QRYTATFVSKNGVGDSVRYGFGSIMWSNAQHQVRSPVAFSWT 993
>TC205344 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (AT4g30020
protein) (AT4g30020/F6G3_50), partial (78%)
Length = 2192
Score = 280 bits (715), Expect = 2e-75
Identities = 217/671 (32%), Positives = 330/671 (48%), Gaps = 47/671 (7%)
Frame = +2
Query: 144 LDTGVWPELKSLDDTGLSPVP--STWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEATLG 201
+D+G++P S P S ++G+CE + S CN K++GA+ F++ A G
Sbjct: 8 VDSGIYPHHPSFTTHNTEPYGPVSRYRGKCEVDPDTKKSFCNGKIVGAQHFAQAAIAA-G 184
Query: 202 PIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCW- 260
+ S D DGHGSHT + AAG + G G A GMA +AR+A YK +
Sbjct: 185 AFNPFIVFDSPLDGDGHGSHTASIAAGRNGIPVRMHGHEFGKASGMAPRARIAVYKALYR 364
Query: 261 LGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTAN-----SHGILVS 315
L G F +D+ A ID+A+ DGV+I+S+S+G +S + F A G+ V+
Sbjct: 365 LFGGFIADVVAAIDQAVHDGVDILSLSVGPNSPPSNTKTTFLNPFDATLLGAVKAGVFVA 544
Query: 316 TSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDS-- 373
+AGNGGP P SL + +PWI TV A DR + ++ LGN G L L+ +
Sbjct: 545 QAAGNGGPFPKSLVSYSPWIATVAAAIDDRRYKNHLILGNGKILAGLGLSPSTRLNQTYT 724
Query: 374 ---PLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERG-----GNARVEK-GLVVK 424
++ + + +S P+ L + + G I++C G+A +++ K
Sbjct: 725 LVAATDVLLDSSVTKYSPTDCQRPELLNKNLIKGNILLCGYSYNFVIGSASIKQVSETAK 904
Query: 425 RAGGIGMIL-ANNEEFGEELVADSHLLPAAALGERS-SKALKDY--VFSSRNPTAKL-VF 479
G +G +L N G + +P + + S SK L DY + + R+ T ++ F
Sbjct: 905 ALGAVGFVLYVENVSPGTKFDPVPVGIPGILITDASKSKELIDYYNISTPRDWTGRVKTF 1084
Query: 480 GGTH--------LQVKPSPVVAAFSSRGPN-----GLTPKILKPDLIAPGVNILAGWTGA 526
GT + K +P VA FS+RGPN +LKPD++APG I A W+
Sbjct: 1085EGTGKIEDGLMPILHKSAPQVAMFSARGPNIKDFSFQEADLLKPDILAPGSLIWAAWS-- 1258
Query: 527 IGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYK 586
+ T P + F +ISGTSM+ PH++G+AA++K HP WSPAAI+SALMTTS T +
Sbjct: 1259LNGTDEP-NYAGEGFAMISGTSMAAPHIAGIAALIKQKHPHWSPAAIKSALMTTSTTLDR 1435
Query: 587 NGQTI-------QDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYT 639
G I + ATP D+G+GHV+P A+LDPGL++DA +DYLGFLC T
Sbjct: 1436AGNPILAQLYSETEAMKLVKATPFDYGSGHVNPRAALDPGLIFDAGYEDYLGFLC----T 1603
Query: 640 SLEIKLASRRDFKCDP--KKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTN 697
+ I + +++ P + N PS + SH + +RT+TN
Sbjct: 1604TPGIDVHEIKNYTNSPCNNTMGHPSNLNTPSITI-----------SHLVRSQIVTRTVTN 1750
Query: 698 VGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKSFAYLYWS 757
V + P+V I V P + + ++ +TVT T S+ +GT SF +
Sbjct: 1751VADEEETYVITARMQPAVAIDVNPPAMTIKASASRR-FTVTLTVRSV-TGTYSFGEVLMK 1924
Query: 758 DGK-HRVASPI 767
+ H+V P+
Sbjct: 1925GSRGHKVRIPV 1957
>TC221447 similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like proteinase,
partial (21%)
Length = 491
Score = 268 bits (686), Expect = 5e-72
Identities = 130/162 (80%), Positives = 141/162 (86%)
Frame = +1
Query: 207 TESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS 266
TES S RDDDGHGSHT TTAAGSAV GASLFG A+GTARGMATQAR+A YKVCWLGGCF+
Sbjct: 1 TESXSPRDDDGHGSHTSTTAAGSAVVGASLFGFANGTARGMATQARLATYKVCWLGGCFT 180
Query: 267 SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPS 326
SDIAAGIDKAIEDGVNI+SMSIGG DY++D IAIG F A +HGILVS SAGNGGPS +
Sbjct: 181 SDIAAGIDKAIEDGVNILSMSIGGGLMDYYKDTIAIGTFAATAHGILVSNSAGNGGPSQA 360
Query: 327 SLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGK 368
+LSN APW+TTVGAGTIDRDFPAYITLGN +TG SLY GK
Sbjct: 361 TLSNVAPWLTTVGAGTIDRDFPAYITLGNGKMYTGVSLYNGK 486
>TC232495 similar to UP|Q8W554 (Q8W554) AT3g14240/MLN21_2, partial (40%)
Length = 731
Score = 263 bits (673), Expect = 2e-70
Identities = 127/233 (54%), Positives = 172/233 (73%), Gaps = 2/233 (0%)
Frame = +3
Query: 440 GEELVADSHLLPAAALGERSSKALKDYVFSSRNP-TAKLVFGGTHLQVKPSPVVAAFSSR 498
GE LVAD H+LPA A+G + ++ Y+ +SR P TA +VF GT L V+P+PVVA+FS+R
Sbjct: 9 GEGLVADCHVLPATAVGATAGDEIRSYIGNSRTPATATIVFKGTRLGVRPAPVVASFSAR 188
Query: 499 GPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLA 558
GPN ++P+ILKPD+IAPG+NILA W +GP+G+P D R FNI+SGTSM+CPHVSGLA
Sbjct: 189 GPNPVSPEILKPDVIAPGLNILAAWPDHVGPSGVPSDGRRTEFNILSGTSMACPHVSGLA 368
Query: 559 AILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDP 618
A+LK +HP+WSPA+IRSALMTT+YT G I D +TG ++ D+GAGHV PV +++P
Sbjct: 369 ALLKAAHPDWSPASIRSALMTTAYTVDNKGDPILDESTGNVSSVFDYGAGHVHPVKAMNP 548
Query: 619 GLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCD-PKKKYRVEDFNYPSFA 670
GLVYD + +DY+ FLC NYT+ I++ +RR+ C K+ + NYPS +
Sbjct: 549 GLVYDISSNDYVNFLCNSNYTTNTIRVITRRNADCSGAKRAGHSGNLNYPSLS 707
>TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1,
partial (40%)
Length = 1392
Score = 261 bits (667), Expect = 8e-70
Identities = 173/456 (37%), Positives = 238/456 (51%), Gaps = 8/456 (1%)
Frame = +2
Query: 322 GPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAG 381
GP SS++ +PWI +V A TI R F + LGN + G S+ L + PLVYAG
Sbjct: 5 GPGLSSITTYSPWILSVAASTIGRKFLTKVQLGNGMVFEGVSI-NTFDLKNKMFPLVYAG 181
Query: 382 NASNFSVGY------LCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILAN 435
+ N + GY C +S+ V GKIV+C+ GNA +K V +G GM+L
Sbjct: 182 DVPNTADGYNSSTSRFCYVNSVDKHLVKGKIVLCD--GNASPKK--VGDLSGAAGMLLGA 349
Query: 436 NEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAF 495
+ ++ LP A + R+ K + Y+ S RN TA + +P + +F
Sbjct: 350 TDVKDAPF---TYALPTAFISLRNFKLIHSYMVSLRNSTATIFRSDEDNDDSQTPFIVSF 520
Query: 496 SSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVS 555
SSRGPN LTP LKPDL APGVNILA W+ + D R V +NI SGTSM+CPHVS
Sbjct: 521 SSRGPNPLTPNTLKPDLAAPGVNILAAWSPVYTISEFKGDKRAVQYNIESGTSMACPHVS 700
Query: 556 GLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVAS 615
AA +K HP WSPA I+SALMTT+ T P +GAG ++P+ +
Sbjct: 701 AAAAYVKSFHPNWSPAMIKSALMTTATPM---------SPTLNPDAEFAYGAGLINPLKA 853
Query: 616 LDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDP-KKKYRVEDFNYPSFAVPLE 674
+PGLVYD + DY+ FLC YT +++ ++ +C KK V D N PS A+ +
Sbjct: 854 ANPGLVYDISEADYVKFLCGEGYTDEMLRVLTKDHSRCSKHAKKEAVYDLNLPSLALYVN 1033
Query: 675 TASGIGGGSHAPITVKYSRTLTNVG-TPGTYKASVSSQSPSVKIAVEPQILRFQELYEKK 733
+S + + RT+TNVG +YKA V S S + I V+P +L F + +KK
Sbjct: 1034VSS---------FSRIFHRTVTNVGLATSSYKAKVVSPS-LIDIQVKPNVLSFTSIGQKK 1183
Query: 734 SYTVTFTSNSMPSGTKSFAYLYWSDGKHRVASPIAI 769
S++V N P A L W DG +V SPI +
Sbjct: 1184SFSVIIEGNVNPDILS--ASLVWDDGTFQVRSPIVV 1285
>BE556066
Length = 425
Score = 238 bits (606), Expect = 1e-62
Identities = 118/138 (85%), Positives = 123/138 (88%)
Frame = +3
Query: 294 DYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITL 353
+Y+RDIIAIG+FTA SHGILVSTSAGNGGPS SLSN APWITTVGAGTIDRDFPAYITL
Sbjct: 9 EYYRDIIAIGSFTATSHGILVSTSAGNGGPSQGSLSNVAPWITTVGAGTIDRDFPAYITL 188
Query: 354 GNNITHTGASLYRGKPLSDSPLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERGG 413
G T+TGASLYRGKPLSDSPLPLVYAGNASN SVGYLCL DSL+P KV GKIVICERGG
Sbjct: 189 GTGKTYTGASLYRGKPLSDSPLPLVYAGNASNSSVGYLCLQDSLIPEKVSGKIVICERGG 368
Query: 414 NARVEKGLVVKRAGGIGM 431
N RVEKGLVVK A G GM
Sbjct: 369 NPRVEKGLVVKLARGAGM 422
>TC216759 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(24%)
Length = 542
Score = 231 bits (589), Expect = 9e-61
Identities = 113/170 (66%), Positives = 134/170 (78%)
Frame = +3
Query: 391 LCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLL 450
LC+ +SL PS V GKIVIC+RG + RV KGLVVK+AGG+GMILAN GE LV D+HLL
Sbjct: 33 LCMENSLDPSMVKGKIVICDRGSSPRVAKGLVVKKAGGVGMILANGISNGEGLVGDAHLL 212
Query: 451 PAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKP 510
PA A+G +K Y+ SS+NPTA L F GT L +KP+PV+A+FS+RGPNGL P+ILKP
Sbjct: 213 PACAVGANEGDLIKKYISSSKNPTATLDFKGTILGIKPAPVIASFSARGPNGLNPEILKP 392
Query: 511 DLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAI 560
DLIAPGVNILA WT A+GPTGL DTR FNI+SGTSM+CPHVSG AA+
Sbjct: 393 DLIAPGVNILAAWTEAVGPTGLDSDTRRTEFNILSGTSMACPHVSGAAAL 542
>TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (21%)
Length = 1106
Score = 214 bits (545), Expect = 1e-55
Identities = 132/313 (42%), Positives = 171/313 (54%), Gaps = 8/313 (2%)
Frame = +2
Query: 467 VFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGA 526
V S+ NP A+++ G T L+ KP+P +A+FSSRGPN + P ILKPD+ APGV+ILA WT
Sbjct: 5 VHSTPNPMAQILPGTTVLETKPAPSMASFSSRGPNIVDPNILKPDITAPGVDILAAWTAE 184
Query: 527 IGPTGLPV-DTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAY 585
GPT + D R V +NI SGTSMSCPHV+ A +LK HP WS AAIRSALMTT+ T
Sbjct: 185 DGPTRMTFNDKRVVKYNIFSGTSMSCPHVAAAAVLLKAIHPTWSTAAIRSALMTTAMTTD 364
Query: 586 KNGQTIQDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKL 645
G + D TG PATP G+GH +P + DPGLVYDA+ YL + C L T
Sbjct: 365 NTGHPLTD-ETGNPATPFAMGSGHFNPKRAADPGLVYDASYMGYLLYTCNLGVTQ----- 526
Query: 646 ASRRDFKCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGT-PGTY 704
+ C PK + NYPS + T RT+TNVG Y
Sbjct: 527 NFNITYNC-PKSFLEPFELNYPSIQI-----------HRLYYTKTIKRTVTNVGRGRSVY 670
Query: 705 KASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKS------FAYLYWSD 758
K S S I P IL+F + +K ++T+T T+N TK F + W+
Sbjct: 671 KFSAVSPK-EYSITATPNILKFNHVGQKINFTITVTANWSQIPTKHGPDKYYFGWYAWTH 847
Query: 759 GKHRVASPIAITW 771
H V SP+A+++
Sbjct: 848 QHHIVRSPVAVSF 886
>TC232007 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(20%)
Length = 669
Score = 202 bits (513), Expect(3) = 4e-55
Identities = 98/179 (54%), Positives = 128/179 (70%)
Frame = +3
Query: 12 ALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQSVSESAEI 71
A L +++F Y A ++T Q K TYI+H+ KS MP +F H W++SSL++VS+SAE+
Sbjct: 51 AFLSVVLFLGLYEAAAEQT-QTHKSTYIVHVAKSEMPESFEHHAVWYESSLKTVSDSAEM 227
Query: 72 LYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKTTTLSP 131
+YTY + HG++TRLT +EA L Q G+L+V PE RYEL TTRTP FLG L K+ L P
Sbjct: 228 IYTYDNAIHGYATRLTAEEARLLQRQTGILAVLPETRYELFTTRTPLFLG-LDKSADLFP 404
Query: 132 GSDKQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGAR 190
S S V++GVLDTGVWPE KS DDTGL PVPSTWKG C+ G N +S+CNRK+I ++
Sbjct: 405 ESSSGSDVIVGVLDTGVWPESKSFDDTGLGPVPSTWKGACQTGTNFTASNCNRKVIRSK 581
Score = 27.3 bits (59), Expect(3) = 4e-55
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = +1
Query: 188 GARFFSKGYEATLGPID 204
GARFF+ G EA LGPI+
Sbjct: 574 GARFFA*GVEAMLGPIN 624
Score = 25.8 bits (55), Expect(3) = 4e-55
Identities = 10/11 (90%), Positives = 10/11 (90%)
Frame = +2
Query: 208 ESRSARDDDGH 218
E RSARDDDGH
Sbjct: 635 EXRSARDDDGH 667
>TC228968 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (20%)
Length = 1008
Score = 211 bits (538), Expect = 7e-55
Identities = 134/302 (44%), Positives = 176/302 (57%), Gaps = 19/302 (6%)
Frame = +3
Query: 227 AGSAVAGASLFG-LASGTARGMATQARVAAYKVCW-LGG--------CFSSDIAAGIDKA 276
AG V AS G A GTA G A AR+A YK CW + G C + D+ ID A
Sbjct: 9 AGRVVPNASAIGGFAKGTALGGAPLARLAIYKACWPIKGKSKHEGNICTNIDMLKAIDDA 188
Query: 277 IEDGVNIISMSIGGSSA-DYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWI 335
I DGV+++S+SIG S+ Y D+IA GA A I+V SAGN GP P +LSN APWI
Sbjct: 189 IGDGVDVLSISIGFSAPISYEEDVIARGALHAVRKNIVVVCSAGNSGPLPQTLSNPAPWI 368
Query: 336 TTVGAGTIDRDFPAYITL------GNNIT--HTGASLYRGKPLSDSPLPLVYAGNASNFS 387
TV A T+DR F A I L G +IT H G S Y D P + + N+
Sbjct: 369 ITVAASTVDRSFHAPIKLNGTIIEGRSITPLHMGNSFYPLVLARDVEHPGLPSNNSG--- 539
Query: 388 VGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEELVADS 447
CL ++L P+K GKIV+C RG R++KGL V+RAGG+G IL NN+ G+++ +D
Sbjct: 540 ---FCLDNTLQPNKARGKIVLCMRGQGERLKKGLEVQRAGGVGFILGNNKLNGKDVPSDP 710
Query: 448 HLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKI 507
H +PA + +S L YV S+ NP A+++ G T L+ KP+P +A+FSSRGPN + P I
Sbjct: 711 HFIPATGVSYENSLKLIQYVHSTPNPMAQILPGTTVLETKPAPSMASFSSRGPNIVDPNI 890
Query: 508 LK 509
LK
Sbjct: 891 LK 896
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,750,079
Number of Sequences: 63676
Number of extensions: 428532
Number of successful extensions: 2501
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 2334
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2350
length of query: 772
length of database: 12,639,632
effective HSP length: 105
effective length of query: 667
effective length of database: 5,953,652
effective search space: 3971085884
effective search space used: 3971085884
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0096b.8


